Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tcf7l1
Z-value: 0.93
Transcription factors associated with Tcf7l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf7l1
|
ENSMUSG00000055799.7 | transcription factor 7 like 1 (T cell specific, HMG box) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf7l1 | mm10_v2_chr6_-_72789240_72789262 | 0.63 | 4.4e-05 | Click! |
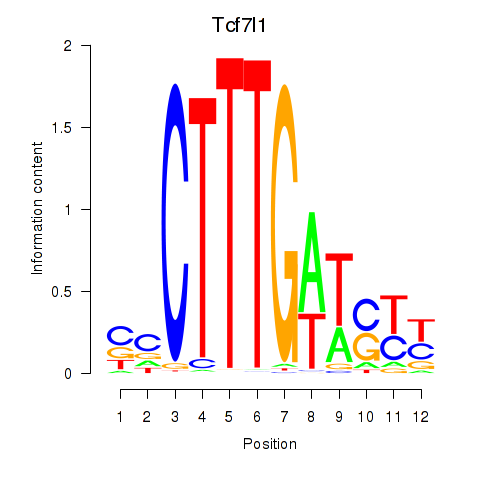
Activity profile of Tcf7l1 motif
Sorted Z-values of Tcf7l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_173021902 | 4.03 |
ENSMUST00000029014.9
|
Rbm38
|
RNA binding motif protein 38 |
| chr12_+_24831583 | 3.74 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr9_+_98490522 | 3.71 |
ENSMUST00000035029.2
|
Rbp2
|
retinol binding protein 2, cellular |
| chr4_+_115057683 | 3.64 |
ENSMUST00000161601.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr18_+_21072329 | 3.64 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr5_-_148392810 | 3.63 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr10_+_43579161 | 3.52 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr2_+_173022360 | 3.41 |
ENSMUST00000173997.1
|
Rbm38
|
RNA binding motif protein 38 |
| chr5_-_122050102 | 3.16 |
ENSMUST00000154139.1
|
Cux2
|
cut-like homeobox 2 |
| chr7_+_45216671 | 2.99 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr2_+_157560078 | 2.98 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr5_+_44100442 | 2.87 |
ENSMUST00000072800.4
|
Gm16401
|
predicted gene 16401 |
| chr6_+_86628174 | 2.84 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr8_+_79028587 | 2.77 |
ENSMUST00000119254.1
|
Zfp827
|
zinc finger protein 827 |
| chr19_+_8591254 | 2.75 |
ENSMUST00000010251.3
ENSMUST00000170817.1 |
Slc22a8
|
solute carrier family 22 (organic anion transporter), member 8 |
| chr1_+_136467958 | 2.46 |
ENSMUST00000047817.6
|
Kif14
|
kinesin family member 14 |
| chr8_-_4259257 | 2.46 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr19_-_41802028 | 2.31 |
ENSMUST00000026150.8
ENSMUST00000177495.1 ENSMUST00000163265.1 |
Arhgap19
|
Rho GTPase activating protein 19 |
| chr11_-_87108656 | 2.26 |
ENSMUST00000051395.8
|
Prr11
|
proline rich 11 |
| chr10_-_116473875 | 2.17 |
ENSMUST00000068233.4
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chrX_-_150812932 | 2.14 |
ENSMUST00000131241.1
ENSMUST00000147152.1 ENSMUST00000143843.1 |
Maged2
|
melanoma antigen, family D, 2 |
| chr4_+_11758147 | 2.13 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chrX_-_150812715 | 2.04 |
ENSMUST00000112697.3
|
Maged2
|
melanoma antigen, family D, 2 |
| chr6_-_83527452 | 1.97 |
ENSMUST00000141904.1
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr7_-_142095266 | 1.93 |
ENSMUST00000039926.3
|
Dusp8
|
dual specificity phosphatase 8 |
| chr8_-_85386310 | 1.91 |
ENSMUST00000137290.1
|
Mylk3
|
myosin light chain kinase 3 |
| chr8_+_79028317 | 1.84 |
ENSMUST00000087927.4
ENSMUST00000098614.2 |
Zfp827
|
zinc finger protein 827 |
| chr1_-_182019927 | 1.77 |
ENSMUST00000078719.6
ENSMUST00000111030.3 ENSMUST00000177811.1 ENSMUST00000111024.3 ENSMUST00000111025.2 |
Enah
|
enabled homolog (Drosophila) |
| chr7_+_100493795 | 1.74 |
ENSMUST00000129324.1
|
Ucp2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr19_-_46327121 | 1.66 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr17_+_34604262 | 1.63 |
ENSMUST00000174041.1
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr2_+_121357714 | 1.62 |
ENSMUST00000125812.1
ENSMUST00000078222.2 ENSMUST00000125221.1 ENSMUST00000150271.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr4_-_43483696 | 1.59 |
ENSMUST00000030180.6
|
Sit1
|
suppression inducing transmembrane adaptor 1 |
| chr7_-_13009795 | 1.54 |
ENSMUST00000051390.7
ENSMUST00000172240.1 |
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr7_-_83884289 | 1.53 |
ENSMUST00000094216.3
|
Mesdc1
|
mesoderm development candidate 1 |
| chr11_+_96929367 | 1.49 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chr5_-_134946917 | 1.49 |
ENSMUST00000051401.2
|
Cldn4
|
claudin 4 |
| chr3_-_50443603 | 1.43 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr4_+_107802277 | 1.40 |
ENSMUST00000106733.2
ENSMUST00000030356.3 ENSMUST00000106732.2 ENSMUST00000126573.1 |
Lrp8
|
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr15_-_98482683 | 1.39 |
ENSMUST00000023726.3
|
Lalba
|
lactalbumin, alpha |
| chr2_+_174760619 | 1.39 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr7_+_98835104 | 1.37 |
ENSMUST00000165122.1
ENSMUST00000067495.2 |
Wnt11
|
wingless-related MMTV integration site 11 |
| chr11_+_46055973 | 1.35 |
ENSMUST00000011400.7
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr8_+_86745679 | 1.32 |
ENSMUST00000098532.2
|
Gm10638
|
predicted gene 10638 |
| chr6_+_17065129 | 1.30 |
ENSMUST00000115467.4
ENSMUST00000154266.2 ENSMUST00000076654.7 |
Tes
|
testis derived transcript |
| chr5_+_37242025 | 1.28 |
ENSMUST00000114158.2
|
Crmp1
|
collapsin response mediator protein 1 |
| chr6_-_52234902 | 1.27 |
ENSMUST00000125581.1
|
Hoxa10
|
homeobox A10 |
| chr1_-_183147461 | 1.26 |
ENSMUST00000171366.1
|
Disp1
|
dispatched homolog 1 (Drosophila) |
| chrX_+_164140447 | 1.24 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr5_-_131538687 | 1.24 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr11_+_67200052 | 1.24 |
ENSMUST00000124516.1
ENSMUST00000018637.8 |
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult |
| chr5_+_33983437 | 1.23 |
ENSMUST00000114384.1
ENSMUST00000094869.5 ENSMUST00000114383.1 |
Gm1673
|
predicted gene 1673 |
| chr9_+_119444923 | 1.20 |
ENSMUST00000035094.6
ENSMUST00000164213.2 |
Exog
|
endo/exonuclease (5'-3'), endonuclease G-like |
| chr2_+_26315513 | 1.19 |
ENSMUST00000066936.2
ENSMUST00000078616.5 |
Gpsm1
|
G-protein signalling modulator 1 (AGS3-like, C. elegans) |
| chr9_-_103761820 | 1.18 |
ENSMUST00000049452.8
|
Tmem108
|
transmembrane protein 108 |
| chr5_+_33983534 | 1.18 |
ENSMUST00000114382.1
|
Gm1673
|
predicted gene 1673 |
| chr11_-_61719946 | 1.17 |
ENSMUST00000151780.1
ENSMUST00000148584.1 |
Slc5a10
|
solute carrier family 5 (sodium/glucose cotransporter), member 10 |
| chr13_+_21945084 | 1.16 |
ENSMUST00000176511.1
ENSMUST00000102978.1 ENSMUST00000152258.2 |
Zfp184
|
zinc finger protein 184 (Kruppel-like) |
| chr12_-_16800674 | 1.16 |
ENSMUST00000162112.1
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr11_+_96929260 | 1.12 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chrX_+_6415736 | 1.11 |
ENSMUST00000143641.3
|
Shroom4
|
shroom family member 4 |
| chr2_+_174760781 | 1.10 |
ENSMUST00000140908.1
|
Edn3
|
endothelin 3 |
| chr14_-_98169542 | 1.09 |
ENSMUST00000069334.7
ENSMUST00000071533.6 |
Dach1
|
dachshund 1 (Drosophila) |
| chr3_+_131110350 | 1.07 |
ENSMUST00000066849.6
ENSMUST00000106341.2 ENSMUST00000029611.7 |
Lef1
|
lymphoid enhancer binding factor 1 |
| chr17_+_12119274 | 1.06 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr12_+_81631369 | 1.06 |
ENSMUST00000036116.5
|
Ttc9
|
tetratricopeptide repeat domain 9 |
| chr5_+_31193227 | 1.06 |
ENSMUST00000031029.8
ENSMUST00000133711.1 ENSMUST00000132471.1 |
Snx17
|
sorting nexin 17 |
| chr19_-_47919269 | 1.06 |
ENSMUST00000095998.5
|
Itprip
|
inositol 1,4,5-triphosphate receptor interacting protein |
| chr4_+_127172866 | 1.06 |
ENSMUST00000106094.2
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3 |
| chr6_-_148944750 | 1.01 |
ENSMUST00000111562.1
ENSMUST00000081956.5 |
Fam60a
|
family with sequence similarity 60, member A |
| chr8_+_70493156 | 0.99 |
ENSMUST00000008032.7
|
Crlf1
|
cytokine receptor-like factor 1 |
| chrX_+_37048807 | 0.98 |
ENSMUST00000060057.1
|
Sowahd
|
sosondowah ankyrin repeat domain family member D |
| chr4_+_141420757 | 0.98 |
ENSMUST00000102486.4
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular) |
| chr17_+_47726834 | 0.97 |
ENSMUST00000024782.5
ENSMUST00000144955.1 |
Pgc
|
progastricsin (pepsinogen C) |
| chrX_-_48034842 | 0.95 |
ENSMUST00000039026.7
|
Apln
|
apelin |
| chr14_+_70774304 | 0.93 |
ENSMUST00000022698.7
|
Dok2
|
docking protein 2 |
| chr11_+_69846610 | 0.93 |
ENSMUST00000152566.1
ENSMUST00000108633.2 |
Plscr3
|
phospholipid scramblase 3 |
| chr9_-_65580040 | 0.93 |
ENSMUST00000068944.7
|
Plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr15_-_98953541 | 0.92 |
ENSMUST00000097014.5
|
Tuba1a
|
tubulin, alpha 1A |
| chr17_+_28530834 | 0.91 |
ENSMUST00000025060.2
|
Armc12
|
armadillo repeat containing 12 |
| chr14_-_70766598 | 0.91 |
ENSMUST00000167242.1
ENSMUST00000022696.6 |
Xpo7
|
exportin 7 |
| chr14_+_51114455 | 0.90 |
ENSMUST00000100645.3
|
Eddm3b
|
epididymal protein 3B |
| chr17_+_7925990 | 0.88 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr6_+_29735667 | 0.88 |
ENSMUST00000001812.4
|
Smo
|
smoothened homolog (Drosophila) |
| chr4_+_127021311 | 0.88 |
ENSMUST00000030623.7
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr14_+_65605267 | 0.87 |
ENSMUST00000079469.6
|
Nuggc
|
nuclear GTPase, germinal center associated |
| chr2_-_35432552 | 0.87 |
ENSMUST00000079424.4
|
Ggta1
|
glycoprotein galactosyltransferase alpha 1, 3 |
| chr13_-_55488038 | 0.86 |
ENSMUST00000109921.2
ENSMUST00000109923.2 ENSMUST00000021950.8 |
Dbn1
|
drebrin 1 |
| chr6_-_122340499 | 0.85 |
ENSMUST00000160843.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr19_-_5894100 | 0.84 |
ENSMUST00000055911.4
|
Tigd3
|
tigger transposable element derived 3 |
| chr11_+_69846665 | 0.84 |
ENSMUST00000019605.2
|
Plscr3
|
phospholipid scramblase 3 |
| chr11_+_5861886 | 0.84 |
ENSMUST00000102923.3
|
Aebp1
|
AE binding protein 1 |
| chr6_-_122340525 | 0.83 |
ENSMUST00000112600.2
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr6_-_52240841 | 0.83 |
ENSMUST00000121043.1
|
Hoxa10
|
homeobox A10 |
| chr19_-_46338632 | 0.82 |
ENSMUST00000051234.8
ENSMUST00000167861.1 |
Cuedc2
|
CUE domain containing 2 |
| chr12_-_84148449 | 0.82 |
ENSMUST00000061425.2
|
Pnma1
|
paraneoplastic antigen MA1 |
| chr2_+_4559742 | 0.79 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr11_-_87074055 | 0.78 |
ENSMUST00000020804.7
|
Gdpd1
|
glycerophosphodiester phosphodiesterase domain containing 1 |
| chr15_-_50889691 | 0.77 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr8_-_69184177 | 0.77 |
ENSMUST00000185176.1
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr6_+_38918969 | 0.77 |
ENSMUST00000003017.6
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr6_+_83057844 | 0.76 |
ENSMUST00000077502.2
|
Dqx1
|
DEAQ RNA-dependent ATPase |
| chr10_+_79988584 | 0.75 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr14_-_70176787 | 0.75 |
ENSMUST00000153735.1
|
Pdlim2
|
PDZ and LIM domain 2 |
| chr11_+_69846374 | 0.74 |
ENSMUST00000108632.1
|
Plscr3
|
phospholipid scramblase 3 |
| chr17_-_51810866 | 0.73 |
ENSMUST00000176669.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr4_-_141599835 | 0.73 |
ENSMUST00000141518.1
ENSMUST00000127455.1 ENSMUST00000105784.1 ENSMUST00000147785.1 |
Fblim1
|
filamin binding LIM protein 1 |
| chr5_-_5266038 | 0.72 |
ENSMUST00000115451.1
ENSMUST00000115452.1 ENSMUST00000131392.1 |
Cdk14
|
cyclin-dependent kinase 14 |
| chr17_-_46202576 | 0.72 |
ENSMUST00000024749.7
|
Polh
|
polymerase (DNA directed), eta (RAD 30 related) |
| chr19_-_32466575 | 0.71 |
ENSMUST00000078034.3
|
Rpl9-ps6
|
ribosomal protein L9, pseudogene 6 |
| chr12_+_31265234 | 0.71 |
ENSMUST00000169088.1
|
Lamb1
|
laminin B1 |
| chr7_-_115824699 | 0.70 |
ENSMUST00000169129.1
|
Sox6
|
SRY-box containing gene 6 |
| chr11_-_54068932 | 0.70 |
ENSMUST00000093109.4
ENSMUST00000018755.3 |
Pdlim4
|
PDZ and LIM domain 4 |
| chr11_+_46404720 | 0.70 |
ENSMUST00000063166.5
|
Fam71b
|
family with sequence similarity 71, member B |
| chr8_-_85067982 | 0.69 |
ENSMUST00000177563.1
|
Gm5741
|
predicted gene 5741 |
| chr11_+_3330781 | 0.69 |
ENSMUST00000136536.1
ENSMUST00000093399.4 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr2_+_163658370 | 0.69 |
ENSMUST00000164399.1
ENSMUST00000064703.6 ENSMUST00000099105.2 ENSMUST00000152418.1 ENSMUST00000126182.1 ENSMUST00000131228.1 |
Pkig
|
protein kinase inhibitor, gamma |
| chr12_+_31265279 | 0.68 |
ENSMUST00000002979.8
ENSMUST00000170495.1 |
Lamb1
|
laminin B1 |
| chr14_+_54936456 | 0.68 |
ENSMUST00000037814.6
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr5_-_134552414 | 0.68 |
ENSMUST00000100647.2
ENSMUST00000036999.6 |
Clip2
|
CAP-GLY domain containing linker protein 2 |
| chr18_-_61036189 | 0.68 |
ENSMUST00000025521.8
|
Cdx1
|
caudal type homeobox 1 |
| chr4_+_107367757 | 0.64 |
ENSMUST00000139560.1
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr4_+_129906205 | 0.63 |
ENSMUST00000145196.1
ENSMUST00000141731.1 ENSMUST00000129149.1 |
E330017L17Rik
|
RIKEN cDNA E330017L17 gene |
| chr9_+_21616166 | 0.62 |
ENSMUST00000034707.8
ENSMUST00000098948.3 |
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr2_+_167932327 | 0.62 |
ENSMUST00000029053.7
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chrX_-_57393020 | 0.62 |
ENSMUST00000143310.1
ENSMUST00000098470.2 ENSMUST00000114726.1 |
Rbmx
|
RNA binding motif protein, X chromosome |
| chr1_+_87470258 | 0.62 |
ENSMUST00000027476.4
|
3110079O15Rik
|
RIKEN cDNA 3110079O15 gene |
| chr17_-_47924460 | 0.61 |
ENSMUST00000113262.1
|
Foxp4
|
forkhead box P4 |
| chr3_-_25212720 | 0.58 |
ENSMUST00000091289.3
|
Gm10259
|
predicted pseudogene 10259 |
| chr8_+_31111816 | 0.58 |
ENSMUST00000046941.7
|
Rnf122
|
ring finger protein 122 |
| chr1_-_36683115 | 0.57 |
ENSMUST00000170295.1
ENSMUST00000114981.1 |
Fam178b
|
family with sequence similarity 178, member B |
| chr10_-_22731336 | 0.56 |
ENSMUST00000127698.1
|
Tbpl1
|
TATA box binding protein-like 1 |
| chr10_-_52195244 | 0.55 |
ENSMUST00000020045.3
|
Ros1
|
Ros1 proto-oncogene |
| chr17_-_47924400 | 0.54 |
ENSMUST00000113263.1
ENSMUST00000097311.2 |
Foxp4
|
forkhead box P4 |
| chrX_-_167209149 | 0.54 |
ENSMUST00000112176.1
|
Tmsb4x
|
thymosin, beta 4, X chromosome |
| chr16_+_19760902 | 0.53 |
ENSMUST00000119468.1
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr14_+_119138415 | 0.53 |
ENSMUST00000065904.3
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr12_+_21417872 | 0.52 |
ENSMUST00000180671.1
|
Gm4419
|
predicted gene 4419 |
| chr3_+_32708546 | 0.52 |
ENSMUST00000029214.7
|
Actl6a
|
actin-like 6A |
| chr1_+_110099295 | 0.52 |
ENSMUST00000134301.1
|
Cdh7
|
cadherin 7, type 2 |
| chr17_+_75005523 | 0.51 |
ENSMUST00000001927.5
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr4_+_132638987 | 0.50 |
ENSMUST00000135299.1
ENSMUST00000081726.6 ENSMUST00000180250.1 ENSMUST00000020197.7 ENSMUST00000079157.4 |
Eya3
|
eyes absent 3 homolog (Drosophila) |
| chr4_-_94650092 | 0.49 |
ENSMUST00000107101.1
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr11_+_3649759 | 0.49 |
ENSMUST00000140242.1
|
Morc2a
|
microrchidia 2A |
| chr12_+_84996309 | 0.49 |
ENSMUST00000168977.1
ENSMUST00000021670.8 |
Ylpm1
|
YLP motif containing 1 |
| chr6_-_47594967 | 0.48 |
ENSMUST00000081721.6
ENSMUST00000114618.1 ENSMUST00000114616.1 |
Ezh2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr13_+_94083490 | 0.48 |
ENSMUST00000156071.1
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2 |
| chr4_-_129742275 | 0.47 |
ENSMUST00000066257.5
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr12_+_53248677 | 0.46 |
ENSMUST00000101432.2
|
Npas3
|
neuronal PAS domain protein 3 |
| chr2_+_157087042 | 0.45 |
ENSMUST00000166140.1
|
Tldc2
|
TBC/LysM associated domain containing 2 |
| chr9_+_35423582 | 0.44 |
ENSMUST00000154652.1
|
Cdon
|
cell adhesion molecule-related/down-regulated by oncogenes |
| chr17_-_35697971 | 0.44 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr4_+_85205120 | 0.43 |
ENSMUST00000107188.3
|
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr17_-_89910449 | 0.43 |
ENSMUST00000086423.4
|
Gm10184
|
predicted pseudogene 10184 |
| chr7_+_73375494 | 0.43 |
ENSMUST00000094312.5
ENSMUST00000119206.1 |
Rgma
|
RGM domain family, member A |
| chr12_-_24096968 | 0.42 |
ENSMUST00000101538.3
|
9030624G23Rik
|
RIKEN cDNA 9030624G23 gene |
| chr8_-_122678072 | 0.42 |
ENSMUST00000006525.7
ENSMUST00000064674.6 |
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr2_+_124610573 | 0.42 |
ENSMUST00000103239.3
ENSMUST00000103240.2 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr5_-_147307264 | 0.42 |
ENSMUST00000031650.3
|
Cdx2
|
caudal type homeobox 2 |
| chr4_+_62583568 | 0.41 |
ENSMUST00000098031.3
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr18_+_82914632 | 0.41 |
ENSMUST00000071233.6
|
Zfp516
|
zinc finger protein 516 |
| chr14_-_101729690 | 0.40 |
ENSMUST00000066461.3
|
Gm9922
|
predicted gene 9922 |
| chr9_+_72925622 | 0.40 |
ENSMUST00000038489.5
|
Pygo1
|
pygopus 1 |
| chr12_+_84996487 | 0.40 |
ENSMUST00000101202.3
|
Ylpm1
|
YLP motif containing 1 |
| chr10_+_53596936 | 0.39 |
ENSMUST00000020004.6
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae) |
| chr12_-_54986363 | 0.39 |
ENSMUST00000173433.1
ENSMUST00000173803.1 |
Baz1a
Gm20403
|
bromodomain adjacent to zinc finger domain 1A predicted gene 20403 |
| chr11_+_96316684 | 0.38 |
ENSMUST00000049241.7
|
Hoxb4
|
homeobox B4 |
| chr9_-_45984816 | 0.37 |
ENSMUST00000172450.1
|
Pafah1b2
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 2 |
| chrX_+_169879596 | 0.37 |
ENSMUST00000112105.1
ENSMUST00000078947.5 |
Mid1
|
midline 1 |
| chr1_-_78968079 | 0.36 |
ENSMUST00000049117.5
|
Gm5830
|
predicted pseudogene 5830 |
| chr10_-_13388830 | 0.35 |
ENSMUST00000079698.5
|
Phactr2
|
phosphatase and actin regulator 2 |
| chrX_-_7572843 | 0.35 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr13_-_97747399 | 0.35 |
ENSMUST00000144993.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr1_-_45925570 | 0.34 |
ENSMUST00000027137.4
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr15_+_102990576 | 0.34 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr4_-_142239356 | 0.33 |
ENSMUST00000036476.3
|
Kazn
|
kazrin, periplakin interacting protein |
| chr6_-_52218686 | 0.33 |
ENSMUST00000134367.2
|
Hoxa7
|
homeobox A7 |
| chr14_-_93888732 | 0.33 |
ENSMUST00000068992.2
|
Pcdh9
|
protocadherin 9 |
| chrX_+_71556874 | 0.32 |
ENSMUST00000123100.1
|
Hmgb3
|
high mobility group box 3 |
| chr3_+_53463666 | 0.32 |
ENSMUST00000058577.4
|
Proser1
|
proline and serine rich 1 |
| chr6_+_6248659 | 0.32 |
ENSMUST00000181633.1
ENSMUST00000176283.1 ENSMUST00000175814.1 ENSMUST00000181192.1 |
Gm20619
|
predicted gene 20619 |
| chr5_-_114380505 | 0.31 |
ENSMUST00000102581.4
|
Kctd10
|
potassium channel tetramerisation domain containing 10 |
| chr17_+_46202740 | 0.31 |
ENSMUST00000087031.5
|
Xpo5
|
exportin 5 |
| chr10_-_13388753 | 0.31 |
ENSMUST00000105546.1
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr7_-_27196138 | 0.31 |
ENSMUST00000122202.1
ENSMUST00000080356.3 |
Snrpa
|
small nuclear ribonucleoprotein polypeptide A |
| chr10_-_52194956 | 0.30 |
ENSMUST00000117992.2
|
Ros1
|
Ros1 proto-oncogene |
| chr2_-_165034770 | 0.29 |
ENSMUST00000122070.1
ENSMUST00000121377.1 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr17_-_68004075 | 0.29 |
ENSMUST00000024840.5
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr6_-_3498297 | 0.28 |
ENSMUST00000183736.1
|
Hepacam2
|
HEPACAM family member 2 |
| chr6_+_83137089 | 0.28 |
ENSMUST00000121093.1
ENSMUST00000087938.4 |
Rtkn
|
rhotekin |
| chr2_-_165034821 | 0.27 |
ENSMUST00000153905.1
ENSMUST00000040381.8 |
Ncoa5
|
nuclear receptor coactivator 5 |
| chr2_+_37516618 | 0.27 |
ENSMUST00000065441.6
|
Gpr21
|
G protein-coupled receptor 21 |
| chr4_+_48585135 | 0.27 |
ENSMUST00000030032.6
|
Tmeff1
|
transmembrane protein with EGF-like and two follistatin-like domains 1 |
| chr2_-_69586021 | 0.27 |
ENSMUST00000100051.2
ENSMUST00000092551.4 ENSMUST00000080953.5 |
Lrp2
|
low density lipoprotein receptor-related protein 2 |
| chr10_+_11609256 | 0.27 |
ENSMUST00000052902.7
|
Gm9797
|
predicted pseudogene 9797 |
| chr13_-_41358990 | 0.27 |
ENSMUST00000163623.1
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chrX_-_57392962 | 0.26 |
ENSMUST00000114730.1
|
Rbmx
|
RNA binding motif protein, X chromosome |
| chr12_-_41485751 | 0.26 |
ENSMUST00000043884.4
|
Lrrn3
|
leucine rich repeat protein 3, neuronal |
| chr1_-_86111970 | 0.26 |
ENSMUST00000027431.6
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr4_+_108479081 | 0.25 |
ENSMUST00000155068.1
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf7l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 1.2 | 3.5 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.9 | 2.7 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.8 | 2.5 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 0.7 | 2.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.6 | 7.4 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.5 | 4.2 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.5 | 3.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.4 | 2.5 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.4 | 1.2 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 1.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 1.4 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 1.4 | GO:0060031 | mediolateral intercalation(GO:0060031) planar cell polarity pathway involved in gastrula mediolateral intercalation(GO:0060775) |
| 0.3 | 1.3 | GO:1904616 | regulation of actin filament binding(GO:1904529) regulation of actin binding(GO:1904616) |
| 0.3 | 1.9 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.3 | 0.9 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.3 | 1.2 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 0.9 | GO:0007494 | midgut development(GO:0007494) |
| 0.3 | 0.9 | GO:0010643 | cell communication by chemical coupling(GO:0010643) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.3 | 1.1 | GO:0071899 | negative regulation of interleukin-13 production(GO:0032696) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 0.7 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.2 | 3.6 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 1.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.2 | 0.9 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.2 | 1.6 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.2 | 1.9 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.2 | 0.8 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 1.0 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.2 | 0.5 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.2 | 0.5 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.2 | 0.6 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.1 | 1.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.9 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.6 | GO:0009212 | dTTP biosynthetic process(GO:0006235) pyrimidine deoxyribonucleoside triphosphate biosynthetic process(GO:0009212) dTTP metabolic process(GO:0046075) |
| 0.1 | 0.7 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 3.2 | GO:0007614 | short-term memory(GO:0007614) |
| 0.1 | 1.4 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.1 | 0.9 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.1 | 0.6 | GO:0007403 | glial cell fate determination(GO:0007403) |
| 0.1 | 0.7 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 1.6 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.3 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 2.7 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 1.2 | GO:1901250 | regulation of lung goblet cell differentiation(GO:1901249) negative regulation of lung goblet cell differentiation(GO:1901250) |
| 0.1 | 1.7 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.1 | 0.8 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.1 | 1.9 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.1 | 1.3 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.5 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.3 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 1.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.1 | 0.9 | GO:0016446 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 3.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 1.5 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.3 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.1 | 0.5 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.7 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.1 | 2.1 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 1.7 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 0.4 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.1 | 0.5 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.6 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.0 | 2.7 | GO:0006509 | membrane protein ectodomain proteolysis(GO:0006509) |
| 0.0 | 0.4 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.2 | GO:0030576 | Cajal body organization(GO:0030576) |
| 0.0 | 0.4 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 1.1 | GO:1990126 | retrograde transport, endosome to plasma membrane(GO:1990126) |
| 0.0 | 0.7 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.4 | GO:0035372 | protein localization to microtubule(GO:0035372) |
| 0.0 | 0.4 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.7 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.8 | GO:0071260 | cellular response to mechanical stimulus(GO:0071260) |
| 0.0 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.2 | GO:0060339 | negative regulation of type I interferon-mediated signaling pathway(GO:0060339) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) positive regulation of cytoplasmic translation(GO:2000767) |
| 0.0 | 3.0 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.8 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 1.5 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 1.6 | GO:0043029 | T cell homeostasis(GO:0043029) |
| 0.0 | 2.5 | GO:0055092 | cholesterol homeostasis(GO:0042632) sterol homeostasis(GO:0055092) |
| 0.0 | 0.0 | GO:0060983 | epicardium-derived cardiac vascular smooth muscle cell differentiation(GO:0060983) |
| 0.0 | 0.3 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.5 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.1 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 0.8 | GO:0034308 | primary alcohol metabolic process(GO:0034308) |
| 0.0 | 0.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.0 | 4.7 | GO:0008654 | phospholipid biosynthetic process(GO:0008654) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.5 | GO:0042711 | maternal behavior(GO:0042711) |
| 0.0 | 0.4 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.2 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.4 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.8 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 0.8 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 2.2 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.0 | 0.3 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.9 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.7 | GO:0016485 | protein processing(GO:0016485) |
| 0.0 | 1.1 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.4 | GO:0009409 | response to cold(GO:0009409) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.6 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 1.0 | 3.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.5 | 1.4 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.0 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.9 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.2 | 0.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.2 | 1.6 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.9 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.7 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 3.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.8 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 0.9 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.1 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.1 | 0.3 | GO:0042272 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 0.6 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.4 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.6 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.9 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.2 | GO:0044094 | host cell nucleus(GO:0042025) host cell nuclear part(GO:0044094) |
| 0.1 | 2.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 2.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 1.0 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.7 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.5 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.6 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.5 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.5 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 1.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 5.7 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.9 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 1.6 | GO:0030496 | midbody(GO:0030496) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 1.1 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.6 | GO:0043204 | perikaryon(GO:0043204) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.6 | 3.7 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.6 | 2.5 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.5 | 1.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.5 | 3.6 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.4 | 1.4 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.3 | 1.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.3 | 2.7 | GO:0015651 | quaternary ammonium group transmembrane transporter activity(GO:0015651) |
| 0.3 | 1.9 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.3 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.3 | 3.7 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.2 | 1.4 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.2 | 3.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.2 | 2.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.2 | 1.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.5 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.2 | 0.7 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.2 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.2 | 1.4 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 3.7 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.9 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.1 | 1.1 | GO:0001075 | transcription factor activity, RNA polymerase II core promoter sequence-specific binding involved in preinitiation complex assembly(GO:0001075) |
| 0.1 | 3.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 2.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.5 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 1.9 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.9 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.4 | GO:0047179 | platelet-activating factor acetyltransferase activity(GO:0047179) |
| 0.1 | 1.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.1 | 2.5 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.5 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 2.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 1.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.3 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.1 | 0.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 1.9 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 7.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 2.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 0.9 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.5 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.1 | 1.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.1 | 0.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 0.3 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.1 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.2 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.1 | 3.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.6 | GO:0001164 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.4 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.7 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.8 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.7 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.9 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.7 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 1.1 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 1.4 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0004910 | interleukin-1, Type II, blocking receptor activity(GO:0004910) |
| 0.0 | 0.7 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 3.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.8 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.6 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.5 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 1.8 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.9 | GO:0003727 | single-stranded RNA binding(GO:0003727) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.4 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 4.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.9 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 1.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 2.0 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 2.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 2.4 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.9 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 3.7 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.1 | 4.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 4.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 2.7 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.1 | 1.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 0.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.1 | 1.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 2.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.9 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.1 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.7 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.5 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 1.4 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.0 | 1.6 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.7 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |