Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
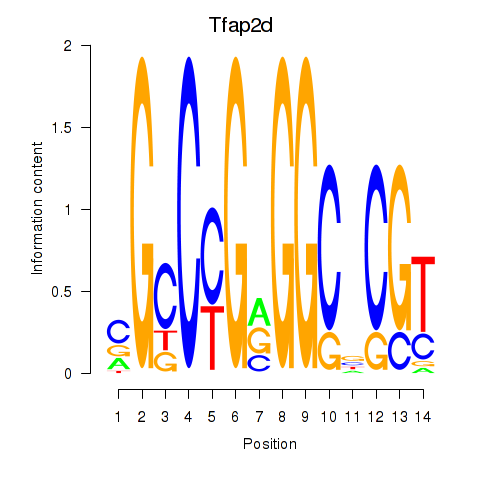
Results for Tfap2d
Z-value: 0.65
Transcription factors associated with Tfap2d
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfap2d
|
ENSMUSG00000042596.7 | transcription factor AP-2, delta |
Activity profile of Tfap2d motif
Sorted Z-values of Tfap2d motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_53213447 | 1.51 |
ENSMUST00000031090.6
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr4_-_57143437 | 1.46 |
ENSMUST00000095076.3
ENSMUST00000030142.3 |
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b |
| chr4_+_129513581 | 1.43 |
ENSMUST00000062356.6
|
Marcksl1
|
MARCKS-like 1 |
| chr17_+_36942910 | 1.26 |
ENSMUST00000040498.5
|
Rnf39
|
ring finger protein 39 |
| chr19_-_4698315 | 1.19 |
ENSMUST00000096325.3
|
Gm960
|
predicted gene 960 |
| chr4_-_114908892 | 1.18 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr7_-_35215248 | 1.13 |
ENSMUST00000118444.1
ENSMUST00000122409.1 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr18_+_64340225 | 1.13 |
ENSMUST00000175965.2
ENSMUST00000115145.3 |
Onecut2
|
one cut domain, family member 2 |
| chr7_+_65862029 | 1.12 |
ENSMUST00000055576.5
ENSMUST00000098391.4 |
Pcsk6
|
proprotein convertase subtilisin/kexin type 6 |
| chr10_-_114801364 | 1.10 |
ENSMUST00000061632.7
|
Trhde
|
TRH-degrading enzyme |
| chr4_-_46991842 | 1.09 |
ENSMUST00000107749.2
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr17_+_36943025 | 1.06 |
ENSMUST00000173072.1
|
Rnf39
|
ring finger protein 39 |
| chr5_+_28165690 | 0.99 |
ENSMUST00000036177.7
|
En2
|
engrailed 2 |
| chr7_-_4789541 | 0.96 |
ENSMUST00000168578.1
|
Tmem238
|
transmembrane protein 238 |
| chr11_+_57801575 | 0.91 |
ENSMUST00000020826.5
|
Sap30l
|
SAP30-like |
| chr2_+_156065738 | 0.89 |
ENSMUST00000137966.1
|
Spag4
|
sperm associated antigen 4 |
| chr8_-_69902712 | 0.84 |
ENSMUST00000180068.1
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr12_+_84009481 | 0.83 |
ENSMUST00000168120.1
|
Acot1
|
acyl-CoA thioesterase 1 |
| chr7_-_84151868 | 0.83 |
ENSMUST00000117085.1
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr5_-_34187670 | 0.81 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr4_-_106464167 | 0.80 |
ENSMUST00000049507.5
|
Pcsk9
|
proprotein convertase subtilisin/kexin type 9 |
| chr10_-_83337440 | 0.80 |
ENSMUST00000126617.1
|
Slc41a2
|
solute carrier family 41, member 2 |
| chr2_+_156065180 | 0.74 |
ENSMUST00000038860.5
|
Spag4
|
sperm associated antigen 4 |
| chr17_+_79614900 | 0.74 |
ENSMUST00000040368.2
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr2_+_167688915 | 0.72 |
ENSMUST00000070642.3
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr17_+_35439155 | 0.72 |
ENSMUST00000071951.6
ENSMUST00000078205.7 ENSMUST00000116598.3 ENSMUST00000076256.7 |
H2-Q7
|
histocompatibility 2, Q region locus 7 |
| chr6_-_23839420 | 0.71 |
ENSMUST00000115358.2
ENSMUST00000163871.2 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr11_+_98386450 | 0.70 |
ENSMUST00000041301.7
|
Pnmt
|
phenylethanolamine-N-methyltransferase |
| chr6_-_23839137 | 0.67 |
ENSMUST00000166458.2
ENSMUST00000142913.2 ENSMUST00000115357.1 ENSMUST00000069074.7 ENSMUST00000115361.2 ENSMUST00000018122.7 ENSMUST00000115355.1 ENSMUST00000115356.2 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr1_+_92831614 | 0.67 |
ENSMUST00000045970.6
|
Gpc1
|
glypican 1 |
| chr4_+_141368116 | 0.66 |
ENSMUST00000006380.4
|
Fam131c
|
family with sequence similarity 131, member C |
| chr13_-_52981027 | 0.66 |
ENSMUST00000071065.7
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr4_+_129058133 | 0.64 |
ENSMUST00000030584.4
ENSMUST00000168461.1 ENSMUST00000152565.1 |
Rnf19b
|
ring finger protein 19B |
| chr1_+_169969409 | 0.63 |
ENSMUST00000180638.1
|
3110045C21Rik
|
RIKEN cDNA 3110045C21 gene |
| chr17_+_24470393 | 0.61 |
ENSMUST00000053024.6
|
Pgp
|
phosphoglycolate phosphatase |
| chr12_+_8771405 | 0.61 |
ENSMUST00000171158.1
|
Sdc1
|
syndecan 1 |
| chr9_-_64341288 | 0.60 |
ENSMUST00000068367.7
|
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr8_-_69902558 | 0.60 |
ENSMUST00000110167.3
|
Ndufa13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr7_-_43489967 | 0.59 |
ENSMUST00000107974.1
|
Iglon5
|
IgLON family member 5 |
| chr5_+_119670658 | 0.59 |
ENSMUST00000079719.4
|
Tbx3
|
T-box 3 |
| chr8_+_123653903 | 0.58 |
ENSMUST00000045487.3
|
Rhou
|
ras homolog gene family, member U |
| chr4_-_41640322 | 0.57 |
ENSMUST00000127306.1
|
Enho
|
energy homeostasis associated |
| chr11_+_97799565 | 0.57 |
ENSMUST00000043843.5
|
Lasp1
|
LIM and SH3 protein 1 |
| chr14_+_57999305 | 0.56 |
ENSMUST00000180534.1
|
3110083C13Rik
|
RIKEN cDNA 3110083C13 gene |
| chr5_-_24601961 | 0.55 |
ENSMUST00000030791.7
|
Smarcd3
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr4_+_152008803 | 0.55 |
ENSMUST00000097773.3
|
Klhl21
|
kelch-like 21 |
| chr15_+_88862178 | 0.55 |
ENSMUST00000042818.9
|
Pim3
|
proviral integration site 3 |
| chr4_-_129239165 | 0.55 |
ENSMUST00000097873.3
|
C77080
|
expressed sequence C77080 |
| chr15_-_79742493 | 0.54 |
ENSMUST00000100439.3
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr4_+_98395891 | 0.54 |
ENSMUST00000107030.2
|
Inadl
|
InaD-like (Drosophila) |
| chr1_+_92910758 | 0.54 |
ENSMUST00000027487.8
|
Rnpepl1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr2_+_27677201 | 0.53 |
ENSMUST00000077257.5
|
Rxra
|
retinoid X receptor alpha |
| chr17_-_79020816 | 0.52 |
ENSMUST00000168887.1
ENSMUST00000119284.1 |
Prkd3
|
protein kinase D3 |
| chr19_+_3851972 | 0.50 |
ENSMUST00000025760.6
|
Chka
|
choline kinase alpha |
| chr8_-_95142477 | 0.50 |
ENSMUST00000034240.7
ENSMUST00000169748.1 |
Kifc3
|
kinesin family member C3 |
| chr7_-_127708886 | 0.50 |
ENSMUST00000061468.8
|
Bcl7c
|
B cell CLL/lymphoma 7C |
| chr15_-_79742518 | 0.49 |
ENSMUST00000089311.4
ENSMUST00000046259.7 |
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr5_+_119670825 | 0.49 |
ENSMUST00000121021.1
|
Tbx3
|
T-box 3 |
| chr9_-_64341145 | 0.48 |
ENSMUST00000120760.1
ENSMUST00000168844.2 |
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chrX_+_94234594 | 0.48 |
ENSMUST00000153900.1
|
Klhl15
|
kelch-like 15 |
| chr9_+_46012810 | 0.47 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr13_-_24937585 | 0.47 |
ENSMUST00000037615.6
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr10_-_127534540 | 0.47 |
ENSMUST00000095266.2
|
Nxph4
|
neurexophilin 4 |
| chr2_-_173276144 | 0.46 |
ENSMUST00000139306.1
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr7_-_19023538 | 0.46 |
ENSMUST00000036018.5
|
Foxa3
|
forkhead box A3 |
| chr19_+_3851797 | 0.46 |
ENSMUST00000072055.6
|
Chka
|
choline kinase alpha |
| chr15_+_89089073 | 0.46 |
ENSMUST00000082439.4
|
Selo
|
selenoprotein O |
| chr2_+_24949747 | 0.45 |
ENSMUST00000028350.3
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr1_+_74391479 | 0.45 |
ENSMUST00000027367.7
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr17_-_24696147 | 0.45 |
ENSMUST00000046839.8
|
Gfer
|
growth factor, erv1 (S. cerevisiae)-like (augmenter of liver regeneration) |
| chr9_+_119402444 | 0.45 |
ENSMUST00000035093.8
ENSMUST00000165044.1 |
Acvr2b
|
activin receptor IIB |
| chr11_+_76202007 | 0.44 |
ENSMUST00000094014.3
|
Fam57a
|
family with sequence similarity 57, member A |
| chr4_-_116017854 | 0.43 |
ENSMUST00000049095.5
|
Faah
|
fatty acid amide hydrolase |
| chr6_+_124996681 | 0.43 |
ENSMUST00000032479.4
|
Pianp
|
PILR alpha associated neural protein |
| chr11_-_72796028 | 0.42 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr5_+_117976761 | 0.42 |
ENSMUST00000035579.6
|
Fbxo21
|
F-box protein 21 |
| chr7_+_64501949 | 0.42 |
ENSMUST00000138829.1
|
Apba2
|
amyloid beta (A4) precursor protein-binding, family A, member 2 |
| chr1_-_74951651 | 0.42 |
ENSMUST00000164097.1
|
Ihh
|
Indian hedgehog |
| chr5_+_30588078 | 0.42 |
ENSMUST00000066295.2
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr19_+_42147373 | 0.42 |
ENSMUST00000061111.9
|
Marveld1
|
MARVEL (membrane-associating) domain containing 1 |
| chr5_-_128433066 | 0.41 |
ENSMUST00000044441.6
|
Tmem132d
|
transmembrane protein 132D |
| chr13_-_43480973 | 0.41 |
ENSMUST00000144326.2
|
Ranbp9
|
RAN binding protein 9 |
| chr10_+_61695503 | 0.41 |
ENSMUST00000020284.4
|
Tysnd1
|
trypsin domain containing 1 |
| chr19_-_5457397 | 0.41 |
ENSMUST00000179549.1
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr17_-_26069409 | 0.40 |
ENSMUST00000120691.1
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II) |
| chr2_+_27677234 | 0.40 |
ENSMUST00000166775.1
|
Rxra
|
retinoid X receptor alpha |
| chr9_-_57645561 | 0.40 |
ENSMUST00000034863.6
|
Csk
|
c-src tyrosine kinase |
| chr8_-_93810225 | 0.39 |
ENSMUST00000181864.1
|
Gm26843
|
predicted gene, 26843 |
| chr5_-_33433976 | 0.39 |
ENSMUST00000173348.1
|
Nkx1-1
|
NK1 transcription factor related, locus 1 (Drosophila) |
| chr11_-_72795801 | 0.38 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr6_+_29526625 | 0.38 |
ENSMUST00000004392.5
|
Irf5
|
interferon regulatory factor 5 |
| chr19_-_4698668 | 0.38 |
ENSMUST00000177696.1
|
Gm960
|
predicted gene 960 |
| chr11_-_102296618 | 0.38 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr11_+_76202084 | 0.38 |
ENSMUST00000169560.1
|
Fam57a
|
family with sequence similarity 57, member A |
| chr7_+_130936172 | 0.37 |
ENSMUST00000006367.7
|
Htra1
|
HtrA serine peptidase 1 |
| chr2_-_30359278 | 0.37 |
ENSMUST00000163668.2
ENSMUST00000028214.8 ENSMUST00000113621.3 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr16_-_46496772 | 0.37 |
ENSMUST00000149901.1
ENSMUST00000096052.2 |
Pvrl3
|
poliovirus receptor-related 3 |
| chr4_-_33189410 | 0.37 |
ENSMUST00000098181.2
|
Pm20d2
|
peptidase M20 domain containing 2 |
| chr7_-_141193934 | 0.37 |
ENSMUST00000026572.4
ENSMUST00000168550.1 ENSMUST00000097957.4 |
Hras
|
Harvey rat sarcoma virus oncogene |
| chr11_+_94328242 | 0.37 |
ENSMUST00000021227.5
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr6_+_72355425 | 0.37 |
ENSMUST00000069695.2
ENSMUST00000132243.1 |
Tmem150a
|
transmembrane protein 150A |
| chr11_+_98795495 | 0.36 |
ENSMUST00000037915.2
|
Msl1
|
male-specific lethal 1 homolog (Drosophila) |
| chr7_-_28379247 | 0.36 |
ENSMUST00000051241.5
|
Zfp36
|
zinc finger protein 36 |
| chr8_-_94012558 | 0.35 |
ENSMUST00000053766.6
|
Amfr
|
autocrine motility factor receptor |
| chr12_+_113098199 | 0.34 |
ENSMUST00000009099.6
ENSMUST00000109723.1 ENSMUST00000109726.1 ENSMUST00000109727.2 ENSMUST00000069690.4 |
Mta1
|
metastasis associated 1 |
| chr10_-_80399389 | 0.34 |
ENSMUST00000105348.1
|
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr7_-_128740471 | 0.34 |
ENSMUST00000119081.1
ENSMUST00000057557.7 |
Mcmbp
|
MCM (minichromosome maintenance deficient) binding protein |
| chr15_-_83555681 | 0.34 |
ENSMUST00000061882.8
|
Mcat
|
malonyl CoA:ACP acyltransferase (mitochondrial) |
| chr8_-_104395765 | 0.33 |
ENSMUST00000179802.1
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr2_-_30359190 | 0.33 |
ENSMUST00000100215.4
ENSMUST00000113620.3 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr5_+_122707546 | 0.33 |
ENSMUST00000081554.6
ENSMUST00000031429.7 ENSMUST00000139631.1 ENSMUST00000142664.1 |
P2rx4
|
purinergic receptor P2X, ligand-gated ion channel 4 |
| chr19_-_7206234 | 0.32 |
ENSMUST00000123594.1
ENSMUST00000025679.4 |
Otub1
|
OTU domain, ubiquitin aldehyde binding 1 |
| chr4_+_153957230 | 0.32 |
ENSMUST00000058393.2
ENSMUST00000105645.2 |
A430005L14Rik
|
RIKEN cDNA A430005L14 gene |
| chr16_+_17070281 | 0.32 |
ENSMUST00000090199.3
|
Ypel1
|
yippee-like 1 (Drosophila) |
| chr9_-_71771535 | 0.31 |
ENSMUST00000122065.1
ENSMUST00000121322.1 ENSMUST00000072899.2 |
Cgnl1
|
cingulin-like 1 |
| chr2_+_91257323 | 0.31 |
ENSMUST00000111349.2
ENSMUST00000131711.1 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr17_+_4994904 | 0.31 |
ENSMUST00000092723.4
ENSMUST00000115797.2 |
Arid1b
|
AT rich interactive domain 1B (SWI-like) |
| chr9_+_107400043 | 0.31 |
ENSMUST00000166799.1
ENSMUST00000170737.1 |
Cacna2d2
|
calcium channel, voltage-dependent, alpha 2/delta subunit 2 |
| chr1_-_182282738 | 0.31 |
ENSMUST00000035295.5
|
Degs1
|
degenerative spermatocyte homolog 1 (Drosophila) |
| chr14_-_47568427 | 0.30 |
ENSMUST00000042988.6
|
Atg14
|
autophagy related 14 |
| chr2_-_173276526 | 0.30 |
ENSMUST00000036248.6
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr11_+_97799772 | 0.30 |
ENSMUST00000129558.1
|
Lasp1
|
LIM and SH3 protein 1 |
| chr7_+_123982799 | 0.30 |
ENSMUST00000106437.1
|
Hs3st4
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 4 |
| chrX_+_73673150 | 0.30 |
ENSMUST00000033752.7
ENSMUST00000114467.2 |
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr2_-_32982007 | 0.30 |
ENSMUST00000028129.7
|
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8 |
| chr11_+_59306920 | 0.30 |
ENSMUST00000000128.3
ENSMUST00000108783.3 |
Wnt9a
|
wingless-type MMTV integration site 9A |
| chr9_+_106477269 | 0.29 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr12_+_71111418 | 0.29 |
ENSMUST00000021482.4
|
Tomm20l
|
translocase of outer mitochondrial membrane 20 homolog (yeast)-like |
| chr19_-_8774431 | 0.29 |
ENSMUST00000010249.5
|
Tmem179b
|
transmembrane protein 179B |
| chr10_-_80399478 | 0.29 |
ENSMUST00000092295.3
ENSMUST00000105349.1 |
Mbd3
|
methyl-CpG binding domain protein 3 |
| chr11_+_50025309 | 0.29 |
ENSMUST00000054684.7
ENSMUST00000102776.4 |
Rnf130
|
ring finger protein 130 |
| chr18_+_24709436 | 0.29 |
ENSMUST00000037097.7
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr17_+_28313513 | 0.28 |
ENSMUST00000114803.1
ENSMUST00000114801.1 ENSMUST00000114804.3 ENSMUST00000088007.4 |
Fance
|
Fanconi anemia, complementation group E |
| chr13_-_92131494 | 0.28 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr10_+_75060885 | 0.28 |
ENSMUST00000164107.1
|
Bcr
|
breakpoint cluster region |
| chr17_-_63499983 | 0.27 |
ENSMUST00000024761.6
|
Fbxl17
|
F-box and leucine-rich repeat protein 17 |
| chr14_+_122475397 | 0.27 |
ENSMUST00000075888.5
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr5_-_124354671 | 0.27 |
ENSMUST00000031341.4
|
Cdk2ap1
|
CDK2 (cyclin-dependent kinase 2)-associated protein 1 |
| chr5_-_34288318 | 0.27 |
ENSMUST00000094868.3
|
Zfyve28
|
zinc finger, FYVE domain containing 28 |
| chr7_-_37772868 | 0.26 |
ENSMUST00000176205.1
|
Zfp536
|
zinc finger protein 536 |
| chr2_+_90847149 | 0.26 |
ENSMUST00000136872.1
|
Mtch2
|
mitochondrial carrier homolog 2 (C. elegans) |
| chr5_+_63812447 | 0.26 |
ENSMUST00000081747.3
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr2_-_168230575 | 0.25 |
ENSMUST00000109193.1
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr9_+_103112072 | 0.25 |
ENSMUST00000035155.6
|
Rab6b
|
RAB6B, member RAS oncogene family |
| chr7_+_4922251 | 0.25 |
ENSMUST00000047309.5
|
Nat14
|
N-acetyltransferase 14 |
| chr18_-_80986578 | 0.25 |
ENSMUST00000057950.7
|
Sall3
|
sal-like 3 (Drosophila) |
| chr17_-_12318660 | 0.24 |
ENSMUST00000089058.5
|
Map3k4
|
mitogen-activated protein kinase kinase kinase 4 |
| chr6_-_52234902 | 0.24 |
ENSMUST00000125581.1
|
Hoxa10
|
homeobox A10 |
| chr12_-_72917760 | 0.24 |
ENSMUST00000110489.2
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr3_+_40950631 | 0.24 |
ENSMUST00000048490.6
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr15_+_34837381 | 0.24 |
ENSMUST00000072868.3
|
Kcns2
|
K+ voltage-gated channel, subfamily S, 2 |
| chr19_+_38481057 | 0.23 |
ENSMUST00000182481.1
|
Plce1
|
phospholipase C, epsilon 1 |
| chr17_+_34647128 | 0.23 |
ENSMUST00000015605.8
ENSMUST00000182587.1 |
Atf6b
|
activating transcription factor 6 beta |
| chr3_-_87930121 | 0.23 |
ENSMUST00000005016.9
|
Rrnad1
|
ribosomal RNA adenine dimethylase domain containing 1 |
| chr8_-_70700070 | 0.23 |
ENSMUST00000116172.1
|
Gm11175
|
predicted gene 11175 |
| chr2_+_168230597 | 0.23 |
ENSMUST00000099071.3
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr7_+_126759601 | 0.23 |
ENSMUST00000050201.4
ENSMUST00000057669.9 |
Mapk3
|
mitogen-activated protein kinase 3 |
| chr7_-_141327700 | 0.22 |
ENSMUST00000080553.7
|
Deaf1
|
deformed epidermal autoregulatory factor 1 (Drosophila) |
| chr8_+_25754492 | 0.22 |
ENSMUST00000167899.1
|
Gm17484
|
predicted gene, 17484 |
| chr15_-_76538728 | 0.22 |
ENSMUST00000023219.7
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr2_+_90847207 | 0.22 |
ENSMUST00000150232.1
ENSMUST00000111467.3 |
Mtch2
|
mitochondrial carrier homolog 2 (C. elegans) |
| chr7_+_25077205 | 0.22 |
ENSMUST00000179556.1
ENSMUST00000053410.9 |
Zfp574
|
zinc finger protein 574 |
| chr17_+_34647187 | 0.22 |
ENSMUST00000173984.1
|
Atf6b
|
activating transcription factor 6 beta |
| chr2_-_32712728 | 0.22 |
ENSMUST00000009699.9
|
Cdk9
|
cyclin-dependent kinase 9 (CDC2-related kinase) |
| chr8_+_83972951 | 0.22 |
ENSMUST00000005606.6
|
Prkaca
|
protein kinase, cAMP dependent, catalytic, alpha |
| chr7_+_141079759 | 0.22 |
ENSMUST00000066873.4
ENSMUST00000163041.1 |
Pkp3
|
plakophilin 3 |
| chr7_-_80340214 | 0.22 |
ENSMUST00000127997.1
ENSMUST00000032748.8 |
Unc45a
|
unc-45 homolog A (C. elegans) |
| chr19_+_8929628 | 0.22 |
ENSMUST00000096241.4
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr5_-_24527276 | 0.21 |
ENSMUST00000088311.4
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr13_+_72628802 | 0.21 |
ENSMUST00000074372.4
|
Irx2
|
Iroquois related homeobox 2 (Drosophila) |
| chr5_+_9266097 | 0.21 |
ENSMUST00000134991.1
ENSMUST00000069538.7 ENSMUST00000115348.2 |
9330182L06Rik
|
RIKEN cDNA 9330182L06 gene |
| chr13_+_54949388 | 0.21 |
ENSMUST00000026994.7
ENSMUST00000109994.2 |
Unc5a
|
unc-5 homolog A (C. elegans) |
| chr11_-_88718165 | 0.21 |
ENSMUST00000107908.1
|
Msi2
|
musashi RNA-binding protein 2 |
| chr4_+_108619925 | 0.21 |
ENSMUST00000030320.6
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr2_+_129800451 | 0.20 |
ENSMUST00000165413.2
ENSMUST00000166282.2 |
Stk35
|
serine/threonine kinase 35 |
| chr2_+_34772089 | 0.20 |
ENSMUST00000028222.6
ENSMUST00000100171.2 |
Hspa5
|
heat shock protein 5 |
| chr7_+_81213567 | 0.20 |
ENSMUST00000026672.7
|
Pde8a
|
phosphodiesterase 8A |
| chr4_-_108780503 | 0.20 |
ENSMUST00000106658.1
|
Zfyve9
|
zinc finger, FYVE domain containing 9 |
| chr18_+_84088077 | 0.20 |
ENSMUST00000060223.2
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr1_-_60566708 | 0.20 |
ENSMUST00000027168.5
ENSMUST00000090293.4 ENSMUST00000140485.1 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr17_-_29347902 | 0.20 |
ENSMUST00000095427.4
ENSMUST00000118366.1 |
Mtch1
|
mitochondrial carrier homolog 1 (C. elegans) |
| chr2_-_28916412 | 0.20 |
ENSMUST00000050776.2
ENSMUST00000113849.1 |
Barhl1
|
BarH-like 1 (Drosophila) |
| chr2_-_151632471 | 0.20 |
ENSMUST00000137936.1
ENSMUST00000146172.1 ENSMUST00000094456.3 ENSMUST00000148755.1 ENSMUST00000109875.1 ENSMUST00000028951.7 ENSMUST00000109877.3 |
Snph
|
syntaphilin |
| chr7_-_114117761 | 0.20 |
ENSMUST00000069449.5
|
Rras2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr6_+_83086365 | 0.19 |
ENSMUST00000041265.2
|
Lbx2
|
ladybird homeobox homolog 2 (Drosophila) |
| chr3_+_94443315 | 0.19 |
ENSMUST00000029786.7
ENSMUST00000098876.3 |
Mrpl9
|
mitochondrial ribosomal protein L9 |
| chr5_-_33629577 | 0.19 |
ENSMUST00000153696.1
ENSMUST00000045329.3 ENSMUST00000065119.8 |
Fam53a
|
family with sequence similarity 53, member A |
| chr3_+_36065979 | 0.19 |
ENSMUST00000011492.8
|
Acad9
|
acyl-Coenzyme A dehydrogenase family, member 9 |
| chr2_+_152293828 | 0.19 |
ENSMUST00000028963.7
ENSMUST00000144252.1 |
Tbc1d20
|
TBC1 domain family, member 20 |
| chr11_-_20112876 | 0.19 |
ENSMUST00000000137.7
|
Actr2
|
ARP2 actin-related protein 2 |
| chr16_+_20672716 | 0.19 |
ENSMUST00000044783.7
ENSMUST00000115463.1 ENSMUST00000142344.1 ENSMUST00000073840.5 ENSMUST00000140576.1 ENSMUST00000115457.1 |
Eif4g1
|
eukaryotic translation initiation factor 4, gamma 1 |
| chr4_+_48279794 | 0.19 |
ENSMUST00000030029.3
|
Invs
|
inversin |
| chr19_-_5560570 | 0.19 |
ENSMUST00000025861.1
|
Ovol1
|
OVO homolog-like 1 (Drosophila) |
| chr1_+_87755870 | 0.19 |
ENSMUST00000144047.1
ENSMUST00000027512.6 ENSMUST00000113186.1 ENSMUST00000113190.2 |
Atg16l1
|
autophagy related 16-like 1 (S. cerevisiae) |
| chr2_-_26206535 | 0.18 |
ENSMUST00000028302.6
|
Lhx3
|
LIM homeobox protein 3 |
| chr14_+_30825580 | 0.18 |
ENSMUST00000006701.5
|
Tmem110
|
transmembrane protein 110 |
| chr2_-_32288022 | 0.18 |
ENSMUST00000183946.1
ENSMUST00000113400.2 ENSMUST00000050410.4 |
Swi5
|
SWI5 recombination repair homolog (yeast) |
| chr1_-_182409020 | 0.18 |
ENSMUST00000097444.1
|
Gm10517
|
predicted gene 10517 |
| chr4_+_41135743 | 0.17 |
ENSMUST00000040008.3
|
Ube2r2
|
ubiquitin-conjugating enzyme E2R 2 |
| chr15_+_59648644 | 0.17 |
ENSMUST00000118228.1
|
Trib1
|
tribbles homolog 1 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfap2d
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0003167 | atrioventricular bundle cell differentiation(GO:0003167) |
| 0.3 | 1.6 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.2 | 1.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.0 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.2 | 0.6 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.8 | GO:0032805 | positive regulation of low-density lipoprotein particle receptor catabolic process(GO:0032805) |
| 0.2 | 0.9 | GO:0045994 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.2 | 1.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.2 | 0.5 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.2 | 1.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.2 | 0.6 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.1 | 0.7 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.1 | 0.4 | GO:0060220 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.1 | 0.6 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 1.0 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.1 | 0.5 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.1 | 0.5 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.1 | 0.3 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.1 | 0.8 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.1 | 1.2 | GO:0097421 | liver regeneration(GO:0097421) |
| 0.1 | 0.3 | GO:0015881 | creatine transport(GO:0015881) |
| 0.1 | 0.3 | GO:0015755 | fructose transport(GO:0015755) |
| 0.1 | 1.1 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.1 | 0.6 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.1 | 0.5 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.1 | 0.3 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 0.5 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 1.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.1 | 0.4 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.2 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.1 | 0.4 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.1 | 0.3 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.1 | GO:0060492 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.1 | 0.2 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.1 | 0.3 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.1 | 0.2 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.1 | 0.2 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.2 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.1 | 0.4 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.1 | 0.5 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.1 | 0.3 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 1.0 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.1 | 0.2 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 0.3 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.1 | 0.5 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.1 | 0.2 | GO:0019230 | proprioception(GO:0019230) sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.1 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.2 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0097360 | inhibition of neuroepithelial cell differentiation(GO:0002085) axial mesoderm formation(GO:0048320) chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.2 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.3 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.6 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.3 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.0 | 0.3 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.0 | 0.5 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:0001806 | type IV hypersensitivity(GO:0001806) |
| 0.0 | 1.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0071374 | cellular response to parathyroid hormone stimulus(GO:0071374) |
| 0.0 | 0.2 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.3 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0034427 | nuclear-transcribed mRNA catabolic process, exonucleolytic, 3'-5'(GO:0034427) |
| 0.0 | 0.1 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.4 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.5 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.5 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:1904173 | negative regulation of RNA interference(GO:1900369) regulation of histone demethylase activity (H3-K4 specific)(GO:1904173) |
| 0.0 | 0.1 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.6 | GO:0072643 | interferon-gamma secretion(GO:0072643) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 1.0 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.1 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.4 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.7 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.3 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.7 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.7 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.4 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.6 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.3 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:1901970 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.5 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.6 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.3 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0051256 | mitotic spindle midzone assembly(GO:0051256) |
| 0.0 | 0.0 | GO:1904954 | canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.2 | 0.7 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 1.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 0.3 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 1.1 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.1 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.4 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.2 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.1 | 0.7 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.5 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 1.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.7 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 1.7 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.2 | GO:0033061 | DNA recombinase mediator complex(GO:0033061) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.4 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 1.6 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.6 | GO:0045271 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:1902710 | GABA receptor complex(GO:1902710) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.3 | 0.8 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.2 | 1.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.2 | 1.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.2 | 0.7 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.2 | 0.6 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.1 | 2.5 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.9 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.4 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.3 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 0.1 | 0.3 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 1.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.6 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.1 | 0.3 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.1 | 1.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.3 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 0.4 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.7 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.9 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.6 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.0 | 0.3 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 1.0 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.6 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 0.5 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.6 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.8 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.3 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 1.1 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.3 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.0 | 0.3 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.3 | GO:0070182 | DNA polymerase binding(GO:0070182) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 1.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.1 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.4 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.2 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.1 | 1.0 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 1.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 1.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.7 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.7 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.5 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |