Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Tfcp2l1
Z-value: 0.55
Transcription factors associated with Tfcp2l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tfcp2l1
|
ENSMUSG00000026380.9 | transcription factor CP2-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tfcp2l1 | mm10_v2_chr1_+_118627943_118627951 | 0.01 | 9.7e-01 | Click! |
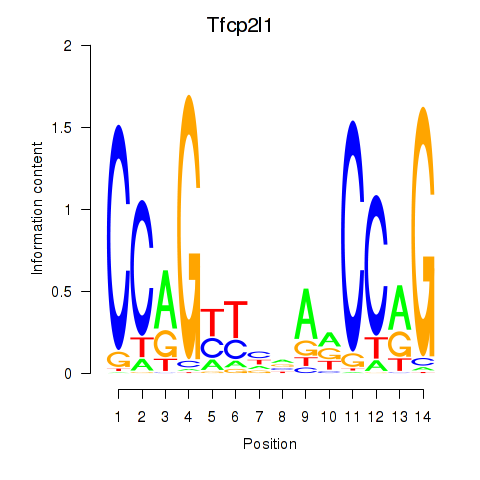
Activity profile of Tfcp2l1 motif
Sorted Z-values of Tfcp2l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_98004634 | 2.19 |
ENSMUST00000131560.1
ENSMUST00000088355.5 |
Col2a1
|
collagen, type II, alpha 1 |
| chr17_-_26199008 | 2.06 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr15_-_98004695 | 2.04 |
ENSMUST00000023123.8
|
Col2a1
|
collagen, type II, alpha 1 |
| chr10_+_43579161 | 1.98 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr16_-_18622403 | 1.48 |
ENSMUST00000167388.1
|
Gp1bb
|
glycoprotein Ib, beta polypeptide |
| chr16_-_22161450 | 1.40 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr14_+_55765956 | 1.40 |
ENSMUST00000057569.3
|
Ltb4r1
|
leukotriene B4 receptor 1 |
| chr8_-_72009619 | 1.35 |
ENSMUST00000003574.4
|
Cyp4f18
|
cytochrome P450, family 4, subfamily f, polypeptide 18 |
| chr7_-_99238564 | 1.22 |
ENSMUST00000064231.7
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr12_-_114406773 | 1.20 |
ENSMUST00000177949.1
|
Ighv6-4
|
immunoglobulin heavy variable V6-4 |
| chr7_-_141117772 | 1.17 |
ENSMUST00000067836.7
|
Ano9
|
anoctamin 9 |
| chr4_-_135573623 | 1.12 |
ENSMUST00000105855.1
|
Grhl3
|
grainyhead-like 3 (Drosophila) |
| chr17_+_35059035 | 1.09 |
ENSMUST00000007255.6
ENSMUST00000174493.1 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr1_+_130731963 | 0.97 |
ENSMUST00000039323.6
|
AA986860
|
expressed sequence AA986860 |
| chr19_+_44759916 | 0.96 |
ENSMUST00000173346.2
|
Pax2
|
paired box gene 2 |
| chr19_+_44757394 | 0.94 |
ENSMUST00000004340.4
|
Pax2
|
paired box gene 2 |
| chr12_+_102128718 | 0.84 |
ENSMUST00000159329.1
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr2_+_121358591 | 0.83 |
ENSMUST00000000317.6
ENSMUST00000129130.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr2_-_91931696 | 0.78 |
ENSMUST00000090602.5
|
Mdk
|
midkine |
| chr3_-_107518001 | 0.77 |
ENSMUST00000169449.1
ENSMUST00000029499.8 |
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr11_-_69602741 | 0.74 |
ENSMUST00000138694.1
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_-_91931774 | 0.73 |
ENSMUST00000069423.6
|
Mdk
|
midkine |
| chr1_+_40681659 | 0.72 |
ENSMUST00000027231.7
|
Slc9a2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 2 |
| chr2_+_91082362 | 0.71 |
ENSMUST00000169852.1
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr7_-_38107490 | 0.69 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr6_-_28831747 | 0.68 |
ENSMUST00000062304.5
|
Lrrc4
|
leucine rich repeat containing 4 |
| chr9_+_32372409 | 0.65 |
ENSMUST00000047334.8
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr2_+_157424255 | 0.63 |
ENSMUST00000029175.7
ENSMUST00000092576.4 |
Src
|
Rous sarcoma oncogene |
| chr2_-_119229849 | 0.61 |
ENSMUST00000110820.2
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr8_-_124434323 | 0.60 |
ENSMUST00000140012.1
|
Pgbd5
|
piggyBac transposable element derived 5 |
| chr4_-_134012381 | 0.60 |
ENSMUST00000176113.1
|
Lin28a
|
lin-28 homolog A (C. elegans) |
| chr8_+_57511833 | 0.59 |
ENSMUST00000067925.6
|
Hmgb2
|
high mobility group box 2 |
| chr2_+_119237351 | 0.58 |
ENSMUST00000028783.7
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr13_-_114932035 | 0.56 |
ENSMUST00000056117.8
|
Itga2
|
integrin alpha 2 |
| chr2_+_119237453 | 0.55 |
ENSMUST00000110816.1
|
Spint1
|
serine protease inhibitor, Kunitz type 1 |
| chr2_-_178414460 | 0.55 |
ENSMUST00000058678.4
|
Ppp1r3d
|
protein phosphatase 1, regulatory subunit 3D |
| chr5_-_138263942 | 0.54 |
ENSMUST00000048421.7
ENSMUST00000164203.1 |
BC037034
BC037034
|
cDNA sequence BC037034 cDNA sequence BC037034 |
| chr4_-_133967893 | 0.52 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_53262547 | 0.51 |
ENSMUST00000098075.2
|
AI427809
|
expressed sequence AI427809 |
| chr18_+_36528145 | 0.50 |
ENSMUST00000074298.6
ENSMUST00000115694.2 |
Slc4a9
|
solute carrier family 4, sodium bicarbonate cotransporter, member 9 |
| chr2_-_119229885 | 0.50 |
ENSMUST00000076084.5
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr9_+_32393963 | 0.49 |
ENSMUST00000172015.1
|
Kcnj1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr16_-_76403673 | 0.47 |
ENSMUST00000052867.7
|
Gm9843
|
predicted gene 9843 |
| chr10_+_80019621 | 0.46 |
ENSMUST00000043311.6
|
Hmha1
|
histocompatibility (minor) HA-1 |
| chr4_-_133967953 | 0.42 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chrX_+_49463926 | 0.41 |
ENSMUST00000130558.1
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr6_-_112946754 | 0.41 |
ENSMUST00000113169.2
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3 |
| chr3_+_95164306 | 0.38 |
ENSMUST00000107217.1
ENSMUST00000168321.1 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr19_-_24477356 | 0.38 |
ENSMUST00000099556.1
|
Fam122a
|
family with sequence similarity 122, member A |
| chr2_+_128126030 | 0.38 |
ENSMUST00000089634.5
ENSMUST00000019281.7 ENSMUST00000110341.2 ENSMUST00000103211.1 ENSMUST00000103210.1 |
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator) |
| chr6_-_136835426 | 0.36 |
ENSMUST00000068293.3
ENSMUST00000111894.1 |
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr12_+_17544873 | 0.32 |
ENSMUST00000171737.1
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr5_-_138264013 | 0.30 |
ENSMUST00000161604.1
ENSMUST00000163268.1 |
BC037034
BC037034
|
cDNA sequence BC037034 cDNA sequence BC037034 |
| chr11_-_69822144 | 0.30 |
ENSMUST00000045771.6
|
Spem1
|
sperm maturation 1 |
| chr14_+_31251454 | 0.30 |
ENSMUST00000022458.4
|
Bap1
|
Brca1 associated protein 1 |
| chr11_+_77982710 | 0.28 |
ENSMUST00000108360.1
ENSMUST00000049167.7 |
Phf12
|
PHD finger protein 12 |
| chr2_-_163750169 | 0.28 |
ENSMUST00000017841.3
|
Ada
|
adenosine deaminase |
| chr2_-_38714491 | 0.27 |
ENSMUST00000028084.4
|
Nr5a1
|
nuclear receptor subfamily 5, group A, member 1 |
| chrX_+_151046131 | 0.26 |
ENSMUST00000112685.1
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr17_+_35194502 | 0.26 |
ENSMUST00000173600.1
|
Ltb
|
lymphotoxin B |
| chr5_+_30105161 | 0.24 |
ENSMUST00000058045.4
|
Gareml
|
GRB2 associated, regulator of MAPK1-like |
| chr6_-_13838432 | 0.24 |
ENSMUST00000115492.1
|
Gpr85
|
G protein-coupled receptor 85 |
| chr3_-_94658800 | 0.23 |
ENSMUST00000107277.1
ENSMUST00000006123.4 ENSMUST00000107279.2 |
Tuft1
|
tuftelin 1 |
| chr3_-_101836223 | 0.23 |
ENSMUST00000061831.4
|
Mab21l3
|
mab-21-like 3 (C. elegans) |
| chr3_+_138860489 | 0.23 |
ENSMUST00000121826.1
|
Tspan5
|
tetraspanin 5 |
| chr11_-_116238077 | 0.22 |
ENSMUST00000037007.3
|
Evpl
|
envoplakin |
| chr17_+_35823509 | 0.22 |
ENSMUST00000173493.1
ENSMUST00000173147.1 ENSMUST00000172846.1 |
Flot1
|
flotillin 1 |
| chr17_+_35823230 | 0.22 |
ENSMUST00000001569.8
ENSMUST00000174080.1 |
Flot1
|
flotillin 1 |
| chr15_-_78718113 | 0.21 |
ENSMUST00000088592.4
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr3_+_89773562 | 0.21 |
ENSMUST00000038356.8
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q (putative) 1 |
| chr8_-_68121527 | 0.21 |
ENSMUST00000178529.1
|
Gm21807
|
predicted gene, 21807 |
| chr9_+_45138437 | 0.20 |
ENSMUST00000060125.5
|
Scn4b
|
sodium channel, type IV, beta |
| chr6_-_129533267 | 0.20 |
ENSMUST00000181594.1
|
1700101I11Rik
|
RIKEN cDNA 1700101I11 gene |
| chr2_+_11339461 | 0.19 |
ENSMUST00000131188.1
|
Gm13293
|
predicted gene 13293 |
| chr17_+_35194405 | 0.19 |
ENSMUST00000025262.5
|
Ltb
|
lymphotoxin B |
| chr4_-_149774238 | 0.19 |
ENSMUST00000105686.2
|
Slc25a33
|
solute carrier family 25, member 33 |
| chr11_+_75531690 | 0.19 |
ENSMUST00000149727.1
ENSMUST00000042561.7 ENSMUST00000108433.1 ENSMUST00000143035.1 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr9_+_37539255 | 0.18 |
ENSMUST00000002008.5
|
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr9_-_108649349 | 0.18 |
ENSMUST00000013338.8
|
Arih2
|
ariadne homolog 2 (Drosophila) |
| chr12_-_11265768 | 0.16 |
ENSMUST00000166117.1
|
Gen1
|
Gen homolog 1, endonuclease (Drosophila) |
| chr14_-_67072465 | 0.14 |
ENSMUST00000089230.5
|
Ppp2r2a
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform |
| chrX_+_151047170 | 0.13 |
ENSMUST00000026296.7
|
Fgd1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr12_-_100520778 | 0.13 |
ENSMUST00000062957.6
|
Ttc7b
|
tetratricopeptide repeat domain 7B |
| chr2_+_156721009 | 0.13 |
ENSMUST00000131157.2
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila) |
| chr15_+_80711292 | 0.12 |
ENSMUST00000067689.7
|
Tnrc6b
|
trinucleotide repeat containing 6b |
| chr11_-_69878018 | 0.12 |
ENSMUST00000178597.1
|
Tmem95
|
transmembrane protein 95 |
| chr4_+_117126776 | 0.12 |
ENSMUST00000134074.1
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr1_+_171329376 | 0.11 |
ENSMUST00000111299.1
ENSMUST00000064950.4 |
Dedd
|
death effector domain-containing |
| chr4_-_116821501 | 0.11 |
ENSMUST00000055436.3
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr19_+_8929628 | 0.11 |
ENSMUST00000096241.4
|
Eml3
|
echinoderm microtubule associated protein like 3 |
| chr4_+_125085134 | 0.11 |
ENSMUST00000154689.1
ENSMUST00000055213.4 ENSMUST00000106171.2 |
Meaf6
|
MYST/Esa1-associated factor 6 |
| chr8_-_17535251 | 0.10 |
ENSMUST00000082104.6
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr16_+_20717665 | 0.10 |
ENSMUST00000021405.7
|
Polr2h
|
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_-_107517321 | 0.10 |
ENSMUST00000166892.1
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chr4_-_106727930 | 0.10 |
ENSMUST00000106770.1
ENSMUST00000145044.1 |
Mroh7
|
maestro heat-like repeat family member 7 |
| chr9_+_44043384 | 0.09 |
ENSMUST00000114840.1
|
Thy1
|
thymus cell antigen 1, theta |
| chrX_-_7319186 | 0.09 |
ENSMUST00000115746.1
|
Clcn5
|
chloride channel 5 |
| chr2_+_156721069 | 0.08 |
ENSMUST00000000094.7
|
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila) |
| chr2_+_156721037 | 0.08 |
ENSMUST00000109566.2
ENSMUST00000146412.2 ENSMUST00000177013.1 ENSMUST00000171030.2 |
Dlgap4
|
discs, large homolog-associated protein 4 (Drosophila) |
| chrX_-_94123087 | 0.08 |
ENSMUST00000113925.1
|
Zfx
|
zinc finger protein X-linked |
| chr11_+_101325063 | 0.08 |
ENSMUST00000041095.7
ENSMUST00000107264.1 |
Aoc2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr9_+_96119362 | 0.08 |
ENSMUST00000085217.5
ENSMUST00000122383.1 |
Gk5
|
glycerol kinase 5 (putative) |
| chr18_+_37806389 | 0.07 |
ENSMUST00000076807.4
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr5_-_35729276 | 0.07 |
ENSMUST00000070203.7
|
Sh3tc1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr11_+_69088490 | 0.07 |
ENSMUST00000021273.6
ENSMUST00000117780.1 |
Vamp2
|
vesicle-associated membrane protein 2 |
| chr5_-_31202215 | 0.06 |
ENSMUST00000176245.1
ENSMUST00000177310.1 ENSMUST00000114590.1 |
Zfp513
|
zinc finger protein 513 |
| chr8_+_123065476 | 0.05 |
ENSMUST00000153285.1
|
Spg7
|
spastic paraplegia 7 homolog (human) |
| chr16_-_18235074 | 0.05 |
ENSMUST00000076957.5
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr15_+_102977032 | 0.05 |
ENSMUST00000001706.6
|
Hoxc9
|
homeobox C9 |
| chr15_-_77970750 | 0.04 |
ENSMUST00000100484.4
|
Eif3d
|
eukaryotic translation initiation factor 3, subunit D |
| chr6_-_113934679 | 0.03 |
ENSMUST00000101044.2
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chr15_+_80234071 | 0.03 |
ENSMUST00000023048.4
ENSMUST00000166030.1 |
Mief1
|
mitochondrial elongation factor 1 |
| chr2_+_91265252 | 0.02 |
ENSMUST00000028691.6
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr3_+_84593547 | 0.02 |
ENSMUST00000062623.3
|
Tigd4
|
tigger transposable element derived 4 |
| chr11_-_109473598 | 0.02 |
ENSMUST00000070152.5
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr5_-_30907692 | 0.01 |
ENSMUST00000132034.2
ENSMUST00000132253.2 |
Ost4
|
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tfcp2l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.6 | 1.9 | GO:0061360 | optic nerve formation(GO:0021634) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.4 | 1.3 | GO:0042374 | menaquinone metabolic process(GO:0009233) phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.4 | 1.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.4 | 1.2 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.3 | 0.6 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.3 | 4.2 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.2 | 1.5 | GO:0030421 | defecation(GO:0030421) |
| 0.2 | 0.7 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.2 | 0.9 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.7 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 1.1 | GO:0070294 | renal sodium ion absorption(GO:0070294) regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.3 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.1 | 0.4 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.3 | GO:0046100 | germinal center B cell differentiation(GO:0002314) adenosine catabolic process(GO:0006154) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) purine nucleobase salvage(GO:0043096) hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) purine deoxyribonucleoside metabolic process(GO:0046122) negative regulation of mucus secretion(GO:0070256) |
| 0.1 | 1.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.1 | 0.4 | GO:0060355 | positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.6 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.6 | GO:0010694 | hypotonic response(GO:0006971) positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.1 | 0.3 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.1 | 1.1 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 1.1 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.7 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 2.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.0 | 0.7 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.5 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.4 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.3 | GO:2000195 | negative regulation of female gonad development(GO:2000195) |
| 0.0 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.2 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.0 | 0.9 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.0 | 1.2 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 1.4 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.7 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0043308 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.2 | GO:0060307 | regulation of ventricular cardiac muscle cell membrane repolarization(GO:0060307) |
| 0.0 | 1.4 | GO:0051028 | mRNA transport(GO:0051028) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.6 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.1 | 2.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.7 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 2.3 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 1.2 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.5 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.6 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.4 | 1.3 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) |
| 0.3 | 1.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.2 | 1.2 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.2 | 1.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.2 | 1.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 2.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.1 | 0.8 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.6 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 1.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.5 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.6 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 0.7 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.1 | 0.6 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 1.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 1.4 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.6 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 1.2 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.9 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 1.3 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.4 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.3 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.6 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 4.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.6 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.7 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 1.1 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 2.1 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |