Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Thra
Z-value: 0.83
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSMUSG00000058756.7 | thyroid hormone receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | mm10_v2_chr11_+_98741805_98741816 | 0.47 | 4.1e-03 | Click! |
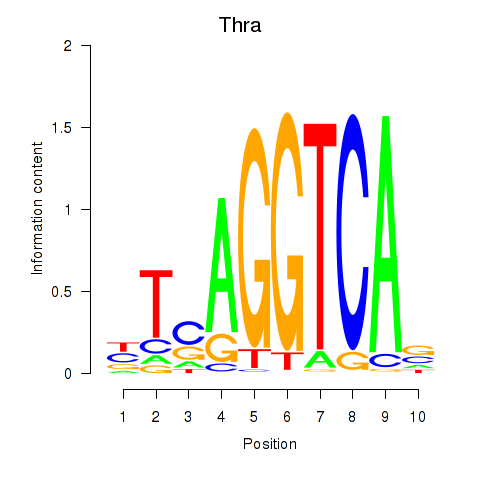
Activity profile of Thra motif
Sorted Z-values of Thra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_7967817 | 5.97 |
ENSMUST00000033502.7
|
Gata1
|
GATA binding protein 1 |
| chr9_-_44288535 | 4.20 |
ENSMUST00000161354.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr9_+_111019284 | 4.02 |
ENSMUST00000035077.3
|
Ltf
|
lactotransferrin |
| chr9_-_44288332 | 3.26 |
ENSMUST00000161408.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chrX_-_102252154 | 2.99 |
ENSMUST00000050551.3
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr2_-_84822546 | 2.84 |
ENSMUST00000028471.5
|
Smtnl1
|
smoothelin-like 1 |
| chr4_+_44300876 | 2.81 |
ENSMUST00000045607.5
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr13_+_95696851 | 2.77 |
ENSMUST00000022182.4
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr11_-_70255329 | 2.74 |
ENSMUST00000108574.2
ENSMUST00000000329.2 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr9_-_44288131 | 2.71 |
ENSMUST00000160384.1
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4 |
| chr2_+_131186942 | 2.66 |
ENSMUST00000028804.8
ENSMUST00000079857.8 |
Cdc25b
|
cell division cycle 25B |
| chr3_+_114030532 | 2.63 |
ENSMUST00000123619.1
ENSMUST00000092155.5 |
Col11a1
|
collagen, type XI, alpha 1 |
| chrX_-_135210672 | 2.50 |
ENSMUST00000033783.1
|
Tceal6
|
transcription elongation factor A (SII)-like 6 |
| chr6_+_86628174 | 2.43 |
ENSMUST00000043400.6
|
Asprv1
|
aspartic peptidase, retroviral-like 1 |
| chr12_+_109545390 | 2.25 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr11_+_69965396 | 2.19 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr13_+_44729794 | 2.13 |
ENSMUST00000172830.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr6_+_113531675 | 2.12 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr10_-_88146867 | 2.06 |
ENSMUST00000164121.1
ENSMUST00000164803.1 ENSMUST00000168163.1 ENSMUST00000048518.9 |
Parpbp
|
PARP1 binding protein |
| chr11_-_33513736 | 2.03 |
ENSMUST00000102815.3
|
Ranbp17
|
RAN binding protein 17 |
| chr15_-_5244164 | 1.83 |
ENSMUST00000120563.1
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_-_5244178 | 1.82 |
ENSMUST00000047379.8
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr15_+_73723131 | 1.73 |
ENSMUST00000165541.1
ENSMUST00000167582.1 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr4_-_156255327 | 1.73 |
ENSMUST00000179919.1
|
Samd11
|
sterile alpha motif domain containing 11 |
| chr3_+_122729158 | 1.72 |
ENSMUST00000066728.5
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr7_+_140093388 | 1.71 |
ENSMUST00000026540.8
|
Prap1
|
proline-rich acidic protein 1 |
| chr7_+_142441808 | 1.71 |
ENSMUST00000105971.1
ENSMUST00000145287.1 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_-_100514800 | 1.60 |
ENSMUST00000054923.7
|
Dnajb13
|
DnaJ (Hsp40) related, subfamily B, member 13 |
| chr9_-_21963568 | 1.57 |
ENSMUST00000006397.5
|
Epor
|
erythropoietin receptor |
| chr11_-_33513626 | 1.53 |
ENSMUST00000037522.7
|
Ranbp17
|
RAN binding protein 17 |
| chr19_-_46044914 | 1.52 |
ENSMUST00000026252.7
|
Ldb1
|
LIM domain binding 1 |
| chr7_+_19411086 | 1.41 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr4_-_140774196 | 1.40 |
ENSMUST00000026381.6
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chrX_+_136666375 | 1.35 |
ENSMUST00000060904.4
ENSMUST00000113100.1 ENSMUST00000128040.1 |
Tceal3
|
transcription elongation factor A (SII)-like 3 |
| chr10_-_62527438 | 1.34 |
ENSMUST00000160987.1
|
Srgn
|
serglycin |
| chr17_+_21555046 | 1.32 |
ENSMUST00000079242.3
|
Zfp52
|
zinc finger protein 52 |
| chr11_-_40733373 | 1.32 |
ENSMUST00000020579.8
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr7_+_35802593 | 1.31 |
ENSMUST00000052454.2
|
E130304I02Rik
|
RIKEN cDNA E130304I02 gene |
| chrX_-_143827391 | 1.30 |
ENSMUST00000087316.5
|
Capn6
|
calpain 6 |
| chrX_+_101449078 | 1.21 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr5_+_110839973 | 1.21 |
ENSMUST00000066160.1
|
Chek2
|
checkpoint kinase 2 |
| chr13_+_44729535 | 1.20 |
ENSMUST00000174068.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr18_+_60774675 | 1.14 |
ENSMUST00000118551.1
|
Rps14
|
ribosomal protein S14 |
| chr4_-_49597860 | 1.14 |
ENSMUST00000042750.2
|
Tmem246
|
transmembrane protein 246 |
| chr16_-_19983005 | 1.13 |
ENSMUST00000058839.8
|
Klhl6
|
kelch-like 6 |
| chrX_-_8145679 | 1.12 |
ENSMUST00000115619.1
ENSMUST00000115617.3 ENSMUST00000040010.3 |
Rbm3
|
RNA binding motif protein 3 |
| chr1_-_84839304 | 1.07 |
ENSMUST00000027421.6
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr11_-_29547820 | 1.07 |
ENSMUST00000102844.3
|
Rps27a
|
ribosomal protein S27A |
| chr18_+_60774510 | 1.07 |
ENSMUST00000025511.3
|
Rps14
|
ribosomal protein S14 |
| chr4_-_49597425 | 1.05 |
ENSMUST00000150664.1
|
Tmem246
|
transmembrane protein 246 |
| chr8_-_105707933 | 1.04 |
ENSMUST00000013299.9
|
Enkd1
|
enkurin domain containing 1 |
| chr1_-_163994767 | 1.03 |
ENSMUST00000097493.3
ENSMUST00000045876.6 |
BC055324
|
cDNA sequence BC055324 |
| chr17_+_80307396 | 1.01 |
ENSMUST00000068175.5
|
Arhgef33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr7_+_28441026 | 0.99 |
ENSMUST00000135686.1
|
Gmfg
|
glia maturation factor, gamma |
| chr10_-_62527413 | 0.98 |
ENSMUST00000160643.1
|
Srgn
|
serglycin |
| chr5_-_134946917 | 0.98 |
ENSMUST00000051401.2
|
Cldn4
|
claudin 4 |
| chr7_+_28440927 | 0.97 |
ENSMUST00000078845.6
|
Gmfg
|
glia maturation factor, gamma |
| chr5_-_110839757 | 0.97 |
ENSMUST00000056937.5
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr3_+_96172327 | 0.96 |
ENSMUST00000076372.4
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr2_+_32525013 | 0.96 |
ENSMUST00000150621.1
|
Gm13412
|
predicted gene 13412 |
| chr8_+_120537423 | 0.93 |
ENSMUST00000118136.1
|
Gse1
|
genetic suppressor element 1 |
| chr13_-_59744921 | 0.91 |
ENSMUST00000180139.1
|
1700014D04Rik
|
RIKEN cDNA 1700014D04 gene |
| chr19_-_5986143 | 0.90 |
ENSMUST00000041827.7
|
Slc22a20
|
solute carrier family 22 (organic anion transporter), member 20 |
| chr15_+_103018936 | 0.82 |
ENSMUST00000165375.1
|
Hoxc4
|
homeobox C4 |
| chr5_-_110839575 | 0.81 |
ENSMUST00000145318.1
|
Hscb
|
HscB iron-sulfur cluster co-chaperone homolog (E. coli) |
| chr14_-_79481268 | 0.81 |
ENSMUST00000022601.5
|
Wbp4
|
WW domain binding protein 4 |
| chr14_-_55106547 | 0.79 |
ENSMUST00000036041.8
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr11_+_85171096 | 0.77 |
ENSMUST00000018623.3
|
1700125H20Rik
|
RIKEN cDNA 1700125H20 gene |
| chr7_+_27258725 | 0.76 |
ENSMUST00000079258.6
|
Numbl
|
numb-like |
| chr6_-_83054415 | 0.75 |
ENSMUST00000113962.1
ENSMUST00000089645.6 ENSMUST00000113963.1 |
Htra2
|
HtrA serine peptidase 2 |
| chr7_+_12922290 | 0.74 |
ENSMUST00000108539.1
ENSMUST00000004554.7 ENSMUST00000147435.1 ENSMUST00000137329.1 |
Rps5
|
ribosomal protein S5 |
| chr19_-_6858179 | 0.72 |
ENSMUST00000113440.1
|
Ccdc88b
|
coiled-coil domain containing 88B |
| chr1_-_167466780 | 0.70 |
ENSMUST00000036643.4
|
Lrrc52
|
leucine rich repeat containing 52 |
| chr17_-_74294834 | 0.70 |
ENSMUST00000078459.6
|
Memo1
|
mediator of cell motility 1 |
| chr2_+_22895482 | 0.69 |
ENSMUST00000053729.7
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chr19_-_46327121 | 0.68 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr1_+_131910458 | 0.68 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chrX_-_57338598 | 0.67 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr11_+_97450136 | 0.67 |
ENSMUST00000107601.1
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr7_+_45514619 | 0.66 |
ENSMUST00000107761.1
|
Tulp2
|
tubby-like protein 2 |
| chr7_+_100472985 | 0.66 |
ENSMUST00000032958.7
ENSMUST00000107059.1 |
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr4_+_156215920 | 0.65 |
ENSMUST00000105572.1
|
2310042D19Rik
|
RIKEN cDNA 2310042D19 gene |
| chr4_-_41048124 | 0.64 |
ENSMUST00000030136.6
|
Aqp7
|
aquaporin 7 |
| chr6_+_120836201 | 0.63 |
ENSMUST00000009256.2
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr2_+_22895583 | 0.61 |
ENSMUST00000152170.1
|
Pdss1
|
prenyl (solanesyl) diphosphate synthase, subunit 1 |
| chrX_-_166585679 | 0.60 |
ENSMUST00000000412.2
|
Egfl6
|
EGF-like-domain, multiple 6 |
| chrX_-_53269020 | 0.60 |
ENSMUST00000114838.1
|
Fam122b
|
family with sequence similarity 122, member B |
| chr3_-_52104891 | 0.60 |
ENSMUST00000121440.1
|
Maml3
|
mastermind like 3 (Drosophila) |
| chr7_+_130865835 | 0.58 |
ENSMUST00000075181.4
ENSMUST00000048180.5 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr3_-_95307132 | 0.58 |
ENSMUST00000015846.2
|
Anxa9
|
annexin A9 |
| chr1_+_160044564 | 0.57 |
ENSMUST00000168359.1
|
4930523C07Rik
|
RIKEN cDNA 4930523C07 gene |
| chr7_-_126976092 | 0.56 |
ENSMUST00000181859.1
|
D830044I16Rik
|
RIKEN cDNA D830044I16 gene |
| chr13_+_54192106 | 0.54 |
ENSMUST00000038101.3
|
Hrh2
|
histamine receptor H2 |
| chr2_+_130406478 | 0.52 |
ENSMUST00000055421.4
|
Tmem239
|
transmembrane 239 |
| chr3_+_95164306 | 0.49 |
ENSMUST00000107217.1
ENSMUST00000168321.1 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr5_-_74065736 | 0.49 |
ENSMUST00000145016.1
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr14_-_118706180 | 0.48 |
ENSMUST00000036554.6
ENSMUST00000166646.1 |
Abcc4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr2_+_112265809 | 0.48 |
ENSMUST00000110991.2
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr7_-_3821812 | 0.48 |
ENSMUST00000153846.1
|
Gm15448
|
predicted gene 15448 |
| chrX_+_145119501 | 0.48 |
ENSMUST00000096301.4
|
Zcchc16
|
zinc finger, CCHC domain containing 16 |
| chr16_-_76403673 | 0.47 |
ENSMUST00000052867.7
|
Gm9843
|
predicted gene 9843 |
| chr11_-_85139939 | 0.47 |
ENSMUST00000108075.2
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr10_-_13324160 | 0.46 |
ENSMUST00000105545.4
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr1_+_152807877 | 0.46 |
ENSMUST00000027754.6
|
Ncf2
|
neutrophil cytosolic factor 2 |
| chr15_+_101412402 | 0.46 |
ENSMUST00000068904.8
|
Krt7
|
keratin 7 |
| chr4_+_33310306 | 0.44 |
ENSMUST00000108153.2
ENSMUST00000029942.7 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr5_-_137625069 | 0.44 |
ENSMUST00000031732.7
|
Fbxo24
|
F-box protein 24 |
| chr17_+_48264270 | 0.44 |
ENSMUST00000059873.7
ENSMUST00000154335.1 ENSMUST00000136272.1 ENSMUST00000125426.1 ENSMUST00000153420.1 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr9_+_44240668 | 0.43 |
ENSMUST00000092426.3
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr4_-_83021102 | 0.43 |
ENSMUST00000071708.5
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr1_-_170306332 | 0.43 |
ENSMUST00000179801.1
|
Gm7694
|
predicted gene 7694 |
| chr14_+_79481164 | 0.42 |
ENSMUST00000040131.5
|
Elf1
|
E74-like factor 1 |
| chr2_+_152911311 | 0.41 |
ENSMUST00000028970.7
|
Mylk2
|
myosin, light polypeptide kinase 2, skeletal muscle |
| chr19_+_8617991 | 0.41 |
ENSMUST00000010250.2
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr7_-_126975400 | 0.39 |
ENSMUST00000180704.1
|
D830044I16Rik
|
RIKEN cDNA D830044I16 gene |
| chr10_-_88379051 | 0.38 |
ENSMUST00000138159.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_+_74542881 | 0.37 |
ENSMUST00000113749.1
ENSMUST00000067916.6 ENSMUST00000113745.1 ENSMUST00000113747.1 ENSMUST00000113750.1 |
Plcd4
|
phospholipase C, delta 4 |
| chr2_+_129592818 | 0.37 |
ENSMUST00000153491.1
ENSMUST00000161620.1 ENSMUST00000179001.1 |
Sirpa
|
signal-regulatory protein alpha |
| chr12_+_78861693 | 0.37 |
ENSMUST00000071230.7
|
Eif2s1
|
eukaryotic translation initiation factor 2, subunit 1 alpha |
| chr9_+_64235201 | 0.36 |
ENSMUST00000039011.3
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr4_+_131873608 | 0.36 |
ENSMUST00000053819.3
|
Srsf4
|
serine/arginine-rich splicing factor 4 |
| chr3_+_95160449 | 0.36 |
ENSMUST00000090823.1
ENSMUST00000090821.3 |
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C |
| chr11_+_40733936 | 0.36 |
ENSMUST00000127382.1
|
Nudcd2
|
NudC domain containing 2 |
| chr16_+_20694908 | 0.34 |
ENSMUST00000056518.6
|
Fam131a
|
family with sequence similarity 131, member A |
| chr14_+_58070547 | 0.34 |
ENSMUST00000165526.1
|
Fgf9
|
fibroblast growth factor 9 |
| chr11_+_31872100 | 0.34 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr11_+_4986824 | 0.33 |
ENSMUST00000009234.9
ENSMUST00000109897.1 |
Ap1b1
|
adaptor protein complex AP-1, beta 1 subunit |
| chr1_+_134193432 | 0.33 |
ENSMUST00000038445.6
|
Mybph
|
myosin binding protein H |
| chr11_+_115475645 | 0.32 |
ENSMUST00000035240.6
|
Armc7
|
armadillo repeat containing 7 |
| chr15_-_78529617 | 0.32 |
ENSMUST00000023075.8
|
C1qtnf6
|
C1q and tumor necrosis factor related protein 6 |
| chr14_+_20929416 | 0.32 |
ENSMUST00000022369.7
|
Vcl
|
vinculin |
| chr10_-_13324250 | 0.32 |
ENSMUST00000105543.1
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr5_+_35057059 | 0.31 |
ENSMUST00000050709.3
|
Dok7
|
docking protein 7 |
| chr10_+_88146992 | 0.31 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr4_-_137766474 | 0.30 |
ENSMUST00000139951.1
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr7_+_45684398 | 0.29 |
ENSMUST00000107742.2
ENSMUST00000183120.1 |
Ntn5
|
netrin 5 |
| chr18_+_9958147 | 0.29 |
ENSMUST00000025137.7
|
Thoc1
|
THO complex 1 |
| chr10_+_88147061 | 0.29 |
ENSMUST00000169309.1
|
Nup37
|
nucleoporin 37 |
| chr5_-_116392766 | 0.28 |
ENSMUST00000153818.1
|
4930562A09Rik
|
RIKEN cDNA 4930562A09 gene |
| chr9_-_99358518 | 0.28 |
ENSMUST00000042158.6
|
Esyt3
|
extended synaptotagmin-like protein 3 |
| chr19_-_37176055 | 0.28 |
ENSMUST00000142973.1
ENSMUST00000154376.1 |
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr12_-_81333129 | 0.27 |
ENSMUST00000085238.6
ENSMUST00000182208.1 |
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr19_+_8764934 | 0.27 |
ENSMUST00000184663.1
|
Nxf1
|
nuclear RNA export factor 1 |
| chr12_-_90738438 | 0.27 |
ENSMUST00000082432.3
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr12_+_86678685 | 0.27 |
ENSMUST00000021681.3
|
Vash1
|
vasohibin 1 |
| chr7_-_4778141 | 0.26 |
ENSMUST00000094892.5
|
Il11
|
interleukin 11 |
| chr17_-_17855188 | 0.25 |
ENSMUST00000003762.6
|
Has1
|
hyaluronan synthase1 |
| chr11_+_54100924 | 0.25 |
ENSMUST00000093107.5
ENSMUST00000019050.5 ENSMUST00000174616.1 ENSMUST00000129499.1 ENSMUST00000126840.1 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr4_-_126202757 | 0.25 |
ENSMUST00000080919.5
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr11_+_40733639 | 0.25 |
ENSMUST00000020578.4
|
Nudcd2
|
NudC domain containing 2 |
| chr5_+_53590215 | 0.24 |
ENSMUST00000037618.6
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr14_+_101653967 | 0.23 |
ENSMUST00000002289.6
|
Uchl3
|
ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase) |
| chr4_+_124880899 | 0.23 |
ENSMUST00000059343.6
|
Epha10
|
Eph receptor A10 |
| chr1_+_106861144 | 0.23 |
ENSMUST00000086701.6
ENSMUST00000112730.1 |
Serpinb5
|
serine (or cysteine) peptidase inhibitor, clade B, member 5 |
| chr12_-_111672290 | 0.23 |
ENSMUST00000001304.7
|
Ckb
|
creatine kinase, brain |
| chr19_-_56389877 | 0.23 |
ENSMUST00000166203.1
ENSMUST00000167239.1 ENSMUST00000040711.8 ENSMUST00000095947.4 ENSMUST00000073536.6 |
Nrap
|
nebulin-related anchoring protein |
| chr4_+_126148457 | 0.23 |
ENSMUST00000106150.2
|
Eva1b
|
eva-1 homolog B (C. elegans) |
| chr4_-_126202583 | 0.23 |
ENSMUST00000106142.1
ENSMUST00000169403.1 ENSMUST00000130334.1 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr19_-_42431778 | 0.21 |
ENSMUST00000048630.6
|
Crtac1
|
cartilage acidic protein 1 |
| chr17_+_35016576 | 0.21 |
ENSMUST00000007245.1
ENSMUST00000172499.1 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr17_-_32388885 | 0.20 |
ENSMUST00000087703.5
ENSMUST00000170603.1 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr15_+_102990576 | 0.20 |
ENSMUST00000001703.6
|
Hoxc8
|
homeobox C8 |
| chr10_+_127195240 | 0.20 |
ENSMUST00000181578.1
|
F420014N23Rik
|
RIKEN cDNA F420014N23 gene |
| chr12_+_79029150 | 0.19 |
ENSMUST00000039928.5
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr6_+_119236507 | 0.19 |
ENSMUST00000037434.6
|
Cacna2d4
|
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr12_+_44328882 | 0.19 |
ENSMUST00000020939.8
ENSMUST00000110748.2 |
Nrcam
|
neuron-glia-CAM-related cell adhesion molecule |
| chr16_+_20696175 | 0.19 |
ENSMUST00000128273.1
|
Fam131a
|
family with sequence similarity 131, member A |
| chr18_+_42275353 | 0.18 |
ENSMUST00000046972.7
ENSMUST00000091920.5 |
Rbm27
|
RNA binding motif protein 27 |
| chr17_-_12992227 | 0.17 |
ENSMUST00000007007.7
|
Wtap
|
Wilms' tumour 1-associating protein |
| chr4_+_94739276 | 0.17 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr1_+_74791516 | 0.17 |
ENSMUST00000006718.8
|
Wnt10a
|
wingless related MMTV integration site 10a |
| chr16_-_26526744 | 0.17 |
ENSMUST00000165687.1
|
Tmem207
|
transmembrane protein 207 |
| chr12_+_102469123 | 0.17 |
ENSMUST00000179218.1
|
Golga5
|
golgi autoantigen, golgin subfamily a, 5 |
| chr10_+_60399726 | 0.17 |
ENSMUST00000164428.1
|
Gm17455
|
predicted gene, 17455 |
| chr7_+_3290553 | 0.17 |
ENSMUST00000096744.5
|
Myadm
|
myeloid-associated differentiation marker |
| chr8_+_72443874 | 0.15 |
ENSMUST00000064853.6
ENSMUST00000121902.1 |
1700030K09Rik
|
RIKEN cDNA 1700030K09 gene |
| chr16_-_31275277 | 0.14 |
ENSMUST00000060188.7
|
Ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2 |
| chr15_+_99702278 | 0.14 |
ENSMUST00000023759.4
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr11_+_7063423 | 0.14 |
ENSMUST00000020706.4
|
Adcy1
|
adenylate cyclase 1 |
| chr7_-_102250086 | 0.13 |
ENSMUST00000106923.1
ENSMUST00000098230.4 |
Rhog
|
ras homolog gene family, member G |
| chr1_+_160195215 | 0.13 |
ENSMUST00000135680.1
ENSMUST00000097193.2 |
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr7_-_105482197 | 0.12 |
ENSMUST00000047040.2
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr4_-_20778527 | 0.10 |
ENSMUST00000119374.1
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr11_+_75650501 | 0.10 |
ENSMUST00000102505.3
|
Myo1c
|
myosin IC |
| chr4_-_141664063 | 0.10 |
ENSMUST00000084203.4
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr6_+_90269106 | 0.10 |
ENSMUST00000058039.2
|
Vmn1r54
|
vomeronasal 1 receptor 54 |
| chr2_-_157279519 | 0.10 |
ENSMUST00000143663.1
|
Mroh8
|
maestro heat-like repeat family member 8 |
| chr14_-_46575847 | 0.10 |
ENSMUST00000151828.1
|
Gm15219
|
predicted gene 15219 |
| chr14_+_32513486 | 0.10 |
ENSMUST00000066807.6
|
Ercc6
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 |
| chr16_-_10790902 | 0.09 |
ENSMUST00000050864.5
|
Prm3
|
protamine 3 |
| chr10_+_126978690 | 0.09 |
ENSMUST00000105256.2
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr10_+_94198955 | 0.09 |
ENSMUST00000020209.9
ENSMUST00000179990.1 |
Ndufa12
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 |
| chr11_+_101984270 | 0.08 |
ENSMUST00000176722.1
ENSMUST00000175972.1 |
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene |
| chr8_+_72107040 | 0.08 |
ENSMUST00000055735.4
|
Olfr374
|
olfactory receptor 374 |
| chr4_+_118409331 | 0.07 |
ENSMUST00000084319.4
ENSMUST00000106384.3 ENSMUST00000126089.1 ENSMUST00000073881.1 ENSMUST00000019229.8 ENSMUST00000144577.1 |
Med8
|
mediator of RNA polymerase II transcription, subunit 8 homolog (yeast) |
| chr11_+_70023905 | 0.07 |
ENSMUST00000124568.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr2_-_25234227 | 0.07 |
ENSMUST00000006638.7
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chr3_-_96172384 | 0.06 |
ENSMUST00000180958.1
|
Gm17690
|
predicted gene, 17690 |
| chr13_+_113351603 | 0.06 |
ENSMUST00000135096.1
|
BC067074
|
cDNA sequence BC067074 |
| chr4_-_135272798 | 0.06 |
ENSMUST00000037099.8
|
Clic4
|
chloride intracellular channel 4 (mitochondrial) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 6.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 1.2 | 3.6 | GO:0032690 | negative regulation of interleukin-1 alpha production(GO:0032690) negative regulation of integrin activation(GO:0033624) negative regulation of interleukin-1 alpha secretion(GO:0050712) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.7 | 4.0 | GO:1902732 | antifungal humoral response(GO:0019732) positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.7 | 2.0 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.6 | 3.0 | GO:0071105 | response to interleukin-9(GO:0071104) response to interleukin-11(GO:0071105) |
| 0.6 | 2.3 | GO:0033370 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.5 | 2.7 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.5 | 1.6 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.4 | 1.7 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.4 | 1.2 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 0.4 | 2.7 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.4 | 1.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.4 | 2.6 | GO:0035989 | tendon development(GO:0035989) |
| 0.4 | 1.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.3 | 1.4 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.3 | 1.8 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.3 | 0.8 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.2 | 0.7 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.2 | 3.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.2 | 2.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.2 | 0.5 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.2 | 2.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.2 | 0.5 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 2.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 1.7 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.1 | 3.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.6 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.1 | 0.5 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.1 | 2.2 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 1.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.6 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.4 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) |
| 0.1 | 0.8 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 6.4 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.1 | 2.8 | GO:0014823 | response to activity(GO:0014823) |
| 0.1 | 0.3 | GO:0002182 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.1 | 0.3 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:0060853 | Notch signaling pathway involved in heart induction(GO:0003137) epidermal cell fate specification(GO:0009957) regulation of Notch signaling pathway involved in heart induction(GO:0035480) positive regulation of Notch signaling pathway involved in heart induction(GO:0035481) arterial endothelial cell fate commitment(GO:0060844) blood vessel endothelial cell fate commitment(GO:0060846) Notch signaling pathway involved in arterial endothelial cell fate commitment(GO:0060853) blood vessel lumenization(GO:0072554) blood vessel endothelial cell fate specification(GO:0097101) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 1.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 2.3 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.1 | 0.4 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.4 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.3 | GO:0048296 | regulation of isotype switching to IgA isotypes(GO:0048296) |
| 0.1 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.3 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.6 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.4 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.1 | 0.3 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.1 | 1.4 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.1 | 0.8 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 2.6 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.8 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 3.6 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.4 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.6 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.5 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 1.4 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.2 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.3 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.7 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.3 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.3 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
| 0.0 | 0.3 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.2 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.2 | 2.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 2.6 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 0.5 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 3.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 3.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.1 | 1.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 4.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 3.2 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 6.1 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.9 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.0 | GO:0005689 | U12-type spliceosomal complex(GO:0005689) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.6 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.3 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 2.5 | GO:0000922 | spindle pole(GO:0000922) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.5 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 1.6 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.1 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.5 | 10.2 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.7 | 2.8 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.7 | 2.7 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.5 | 3.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.4 | 6.0 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.4 | 3.6 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.3 | 1.3 | GO:0050347 | trans-hexaprenyltranstransferase activity(GO:0000010) trans-octaprenyltranstransferase activity(GO:0050347) |
| 0.3 | 1.7 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.3 | 1.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 1.6 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.2 | 2.2 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.2 | 3.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.2 | 2.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.2 | 1.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 0.5 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.2 | 2.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.4 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 4.0 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 0.6 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 1.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.7 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 1.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 2.4 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 3.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.5 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 3.3 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.1 | 2.1 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.1 | 0.4 | GO:0032027 | myosin light chain kinase activity(GO:0004687) myosin light chain binding(GO:0032027) |
| 0.1 | 1.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.3 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.5 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 1.9 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 1.6 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.0 | 0.3 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.9 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.6 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.6 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 2.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 1.3 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 3.9 | GO:0004725 | protein tyrosine phosphatase activity(GO:0004725) |
| 0.0 | 2.3 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.6 | GO:0015464 | acetylcholine receptor activity(GO:0015464) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.8 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.5 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 1.4 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.6 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.4 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.7 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.1 | 2.8 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.1 | 3.3 | PID ATM PATHWAY | ATM pathway |
| 0.1 | 5.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.1 | 1.7 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.1 | 2.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 2.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.8 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 3.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.3 | 3.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.3 | 3.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.2 | 2.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.2 | 1.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 4.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.1 | 3.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 3.3 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 2.8 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.8 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 5.3 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 2.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 1.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.1 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 1.4 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.9 | REACTOME TRANSPORT OF MATURE MRNA DERIVED FROM AN INTRONLESS TRANSCRIPT | Genes involved in Transport of Mature mRNA Derived from an Intronless Transcript |
| 0.0 | 1.0 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.4 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |