Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
Results for Vsx2_Dlx3
Z-value: 1.50
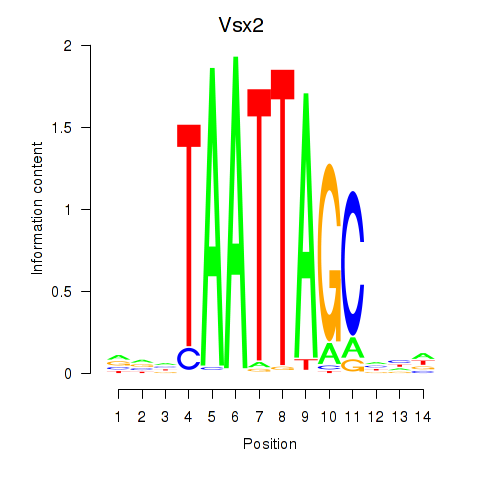
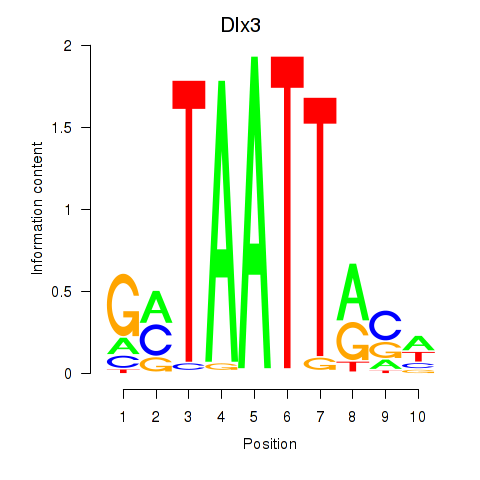
Transcription factors associated with Vsx2_Dlx3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vsx2
|
ENSMUSG00000021239.6 | visual system homeobox 2 |
|
Dlx3
|
ENSMUSG00000001510.7 | distal-less homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Dlx3 | mm10_v2_chr11_+_95120089_95120125 | 0.55 | 5.1e-04 | Click! |
| Vsx2 | mm10_v2_chr12_+_84569762_84569840 | 0.22 | 1.9e-01 | Click! |
Activity profile of Vsx2_Dlx3 motif
Sorted Z-values of Vsx2_Dlx3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_168767136 | 12.44 |
ENSMUST00000029061.5
ENSMUST00000103074.1 |
Sall4
|
sal-like 4 (Drosophila) |
| chr2_-_168767029 | 9.87 |
ENSMUST00000075044.3
|
Sall4
|
sal-like 4 (Drosophila) |
| chr4_+_34893772 | 9.18 |
ENSMUST00000029975.3
ENSMUST00000135871.1 ENSMUST00000108130.1 |
Cga
|
glycoprotein hormones, alpha subunit |
| chr12_+_109545390 | 7.71 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr14_+_27000362 | 7.08 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr2_+_84734050 | 6.39 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr16_-_42340595 | 5.94 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr3_-_116253467 | 5.53 |
ENSMUST00000090473.5
|
Gpr88
|
G-protein coupled receptor 88 |
| chr3_+_51661167 | 5.41 |
ENSMUST00000099106.3
|
Mgst2
|
microsomal glutathione S-transferase 2 |
| chr19_+_38132767 | 5.20 |
ENSMUST00000025956.5
ENSMUST00000112329.1 |
Pde6c
|
phosphodiesterase 6C, cGMP specific, cone, alpha prime |
| chr6_+_30541582 | 4.83 |
ENSMUST00000096066.4
|
Cpa2
|
carboxypeptidase A2, pancreatic |
| chr17_-_28560704 | 4.52 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr10_+_75564086 | 4.24 |
ENSMUST00000141062.1
ENSMUST00000152657.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr7_-_100855403 | 3.47 |
ENSMUST00000156855.1
|
Relt
|
RELT tumor necrosis factor receptor |
| chr14_+_99298652 | 3.20 |
ENSMUST00000005279.6
|
Klf5
|
Kruppel-like factor 5 |
| chr4_-_99654983 | 3.15 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr11_+_62077018 | 2.82 |
ENSMUST00000092415.5
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_-_79285502 | 2.80 |
ENSMUST00000165408.1
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr1_+_172555932 | 2.62 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr1_-_79440039 | 2.60 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr10_-_6980376 | 2.17 |
ENSMUST00000105617.1
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr14_+_32321987 | 2.14 |
ENSMUST00000022480.7
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
| chr3_-_130730375 | 2.13 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr4_+_5724304 | 1.97 |
ENSMUST00000108380.1
|
Fam110b
|
family with sequence similarity 110, member B |
| chr12_+_108605757 | 1.83 |
ENSMUST00000109854.2
|
Evl
|
Ena-vasodilator stimulated phosphoprotein |
| chr6_-_102464667 | 1.78 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr2_-_121235689 | 1.71 |
ENSMUST00000142400.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr12_+_38780284 | 1.70 |
ENSMUST00000162563.1
ENSMUST00000161164.1 ENSMUST00000160996.1 |
Etv1
|
ets variant gene 1 |
| chr17_+_35533194 | 1.64 |
ENSMUST00000025273.8
|
Psors1c2
|
psoriasis susceptibility 1 candidate 2 (human) |
| chr3_+_159839729 | 1.63 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr4_-_132510493 | 1.57 |
ENSMUST00000030724.8
|
Sesn2
|
sestrin 2 |
| chr7_-_115846080 | 1.43 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr9_+_119063429 | 1.41 |
ENSMUST00000141185.1
ENSMUST00000126251.1 ENSMUST00000136561.1 |
Vill
|
villin-like |
| chr15_-_34356421 | 1.37 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr7_+_126950518 | 1.37 |
ENSMUST00000106335.1
ENSMUST00000146017.1 |
Sez6l2
|
seizure related 6 homolog like 2 |
| chr15_+_81936753 | 1.36 |
ENSMUST00000038757.7
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr3_-_130730310 | 1.35 |
ENSMUST00000062601.7
|
Rpl34
|
ribosomal protein L34 |
| chr4_-_41464816 | 1.34 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr6_-_136781718 | 1.29 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr15_+_81936911 | 1.28 |
ENSMUST00000135663.1
|
Csdc2
|
cold shock domain containing C2, RNA binding |
| chr2_+_69219971 | 1.28 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr2_+_127909058 | 1.28 |
ENSMUST00000110344.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr16_-_89818338 | 1.25 |
ENSMUST00000164263.2
|
Tiam1
|
T cell lymphoma invasion and metastasis 1 |
| chr3_-_17230976 | 1.25 |
ENSMUST00000177874.1
|
Gm5283
|
predicted gene 5283 |
| chr7_-_116031047 | 1.24 |
ENSMUST00000106612.1
|
Sox6
|
SRY-box containing gene 6 |
| chr7_+_144838590 | 1.23 |
ENSMUST00000105898.1
|
Fgf3
|
fibroblast growth factor 3 |
| chr19_-_47692042 | 1.23 |
ENSMUST00000026045.7
ENSMUST00000086923.5 |
Col17a1
|
collagen, type XVII, alpha 1 |
| chr1_-_172027251 | 1.22 |
ENSMUST00000138714.1
|
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr17_-_67950908 | 1.21 |
ENSMUST00000164647.1
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr7_+_126950687 | 1.19 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr12_+_38780817 | 1.14 |
ENSMUST00000160856.1
|
Etv1
|
ets variant gene 1 |
| chr2_-_116067391 | 1.11 |
ENSMUST00000140185.1
|
2700033N17Rik
|
RIKEN cDNA 2700033N17 gene |
| chr5_-_122989260 | 1.11 |
ENSMUST00000118027.1
|
Kdm2b
|
lysine (K)-specific demethylase 2B |
| chr15_-_101694299 | 1.05 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr17_-_24073479 | 1.03 |
ENSMUST00000017090.5
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr1_-_172027269 | 1.03 |
ENSMUST00000027837.6
ENSMUST00000111264.1 |
Vangl2
|
vang-like 2 (van gogh, Drosophila) |
| chr5_+_27261916 | 1.00 |
ENSMUST00000101471.3
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr9_+_53771499 | 0.97 |
ENSMUST00000048670.8
|
Slc35f2
|
solute carrier family 35, member F2 |
| chr4_+_134068429 | 0.97 |
ENSMUST00000121391.1
|
Aim1l
|
absent in melanoma 1-like |
| chr11_-_106301801 | 0.94 |
ENSMUST00000103071.3
|
Gh
|
growth hormone |
| chr11_+_29373618 | 0.92 |
ENSMUST00000040182.6
ENSMUST00000109477.1 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr12_+_38781093 | 0.92 |
ENSMUST00000161513.1
|
Etv1
|
ets variant gene 1 |
| chr18_+_37518341 | 0.92 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chrX_+_112093496 | 0.91 |
ENSMUST00000130247.2
ENSMUST00000038546.6 |
Tex16
|
testis expressed gene 16 |
| chrX_+_140956892 | 0.89 |
ENSMUST00000112971.1
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr8_+_57320975 | 0.89 |
ENSMUST00000040104.3
|
Hand2
|
heart and neural crest derivatives expressed transcript 2 |
| chr7_+_126950837 | 0.89 |
ENSMUST00000106332.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr19_-_5560570 | 0.89 |
ENSMUST00000025861.1
|
Ovol1
|
OVO homolog-like 1 (Drosophila) |
| chr2_+_49787675 | 0.88 |
ENSMUST00000028103.6
|
Lypd6b
|
LY6/PLAUR domain containing 6B |
| chr3_-_50443603 | 0.84 |
ENSMUST00000029297.4
|
Slc7a11
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 11 |
| chr2_-_7395879 | 0.81 |
ENSMUST00000182404.1
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr3_-_49757257 | 0.80 |
ENSMUST00000035931.7
|
Pcdh18
|
protocadherin 18 |
| chr9_-_120068263 | 0.79 |
ENSMUST00000064165.3
ENSMUST00000177637.1 |
Cx3cr1
|
chemokine (C-X3-C) receptor 1 |
| chr9_-_21312255 | 0.77 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr4_-_83052147 | 0.74 |
ENSMUST00000170248.2
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr4_-_83052229 | 0.73 |
ENSMUST00000107230.1
|
Frem1
|
Fras1 related extracellular matrix protein 1 |
| chr7_+_66839752 | 0.71 |
ENSMUST00000107478.1
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr2_-_59948155 | 0.70 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr16_+_34690548 | 0.70 |
ENSMUST00000023532.6
|
Ccdc14
|
coiled-coil domain containing 14 |
| chr10_+_69787431 | 0.68 |
ENSMUST00000183240.1
|
Ank3
|
ankyrin 3, epithelial |
| chr15_+_98571004 | 0.67 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr18_+_23415400 | 0.66 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr12_+_99964499 | 0.65 |
ENSMUST00000177549.1
ENSMUST00000160413.1 ENSMUST00000162221.1 ENSMUST00000049788.8 |
Kcnk13
|
potassium channel, subfamily K, member 13 |
| chr7_+_66839726 | 0.65 |
ENSMUST00000098382.3
|
Adamts17
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 17 |
| chr11_+_101987050 | 0.64 |
ENSMUST00000010985.7
|
1700006E09Rik
|
RIKEN cDNA 1700006E09 gene |
| chr9_+_18427543 | 0.62 |
ENSMUST00000053326.9
|
Gm5612
|
predicted gene 5612 |
| chrX_+_106583184 | 0.61 |
ENSMUST00000101296.2
ENSMUST00000101297.3 |
Gm5127
|
predicted gene 5127 |
| chr14_+_76504478 | 0.61 |
ENSMUST00000022587.9
ENSMUST00000134109.1 |
Tsc22d1
|
TSC22 domain family, member 1 |
| chr12_+_52699297 | 0.61 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr18_+_82914632 | 0.60 |
ENSMUST00000071233.6
|
Zfp516
|
zinc finger protein 516 |
| chr17_+_17402672 | 0.57 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr1_-_134955908 | 0.57 |
ENSMUST00000045665.6
ENSMUST00000086444.4 ENSMUST00000112163.1 |
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr10_-_28986280 | 0.54 |
ENSMUST00000152363.1
ENSMUST00000015663.6 |
2310057J18Rik
|
RIKEN cDNA 2310057J18 gene |
| chr5_-_115652974 | 0.52 |
ENSMUST00000121746.1
ENSMUST00000118576.1 |
Ccdc64
|
coiled-coil domain containing 64 |
| chr2_+_153938218 | 0.52 |
ENSMUST00000109757.1
|
Bpifb4
|
BPI fold containing family B, member 4 |
| chr11_-_71144704 | 0.50 |
ENSMUST00000108518.2
|
Nlrp1a
|
NLR family, pyrin domain containing 1A |
| chr17_-_35697971 | 0.50 |
ENSMUST00000146472.1
|
Ddr1
|
discoidin domain receptor family, member 1 |
| chr18_+_44104407 | 0.49 |
ENSMUST00000081271.5
|
Spink12
|
serine peptidase inhibitor, Kazal type 11 |
| chr14_+_76504185 | 0.48 |
ENSMUST00000177207.1
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr4_+_8690399 | 0.48 |
ENSMUST00000127476.1
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr2_+_127854628 | 0.47 |
ENSMUST00000028859.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chrX_+_164162167 | 0.47 |
ENSMUST00000131543.1
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr1_-_134955847 | 0.45 |
ENSMUST00000168381.1
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B |
| chr17_-_70998010 | 0.45 |
ENSMUST00000024846.6
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr8_-_3624989 | 0.44 |
ENSMUST00000142431.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr6_-_116716888 | 0.44 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr5_-_104077608 | 0.41 |
ENSMUST00000164471.1
ENSMUST00000178967.1 |
Gm17660
|
predicted gene, 17660 |
| chr5_+_14025305 | 0.41 |
ENSMUST00000073957.6
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr8_-_57653023 | 0.41 |
ENSMUST00000034021.5
|
Galnt7
|
UDP-N-acetyl-alpha-D-galactosamine: polypeptide N-acetylgalactosaminyltransferase 7 |
| chr2_+_36230426 | 0.41 |
ENSMUST00000062069.5
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr13_-_102906046 | 0.39 |
ENSMUST00000171791.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr13_+_118714678 | 0.37 |
ENSMUST00000022246.8
|
Fgf10
|
fibroblast growth factor 10 |
| chr8_-_120228221 | 0.33 |
ENSMUST00000183235.1
|
A330074K22Rik
|
RIKEN cDNA A330074K22 gene |
| chr9_-_37147257 | 0.33 |
ENSMUST00000039674.5
ENSMUST00000080754.5 |
Pknox2
|
Pbx/knotted 1 homeobox 2 |
| chr8_-_34965631 | 0.33 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr4_+_62583568 | 0.33 |
ENSMUST00000098031.3
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr4_+_110397764 | 0.31 |
ENSMUST00000097920.2
ENSMUST00000080744.6 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr9_-_79977782 | 0.31 |
ENSMUST00000093811.4
|
Filip1
|
filamin A interacting protein 1 |
| chr8_-_3625274 | 0.29 |
ENSMUST00000004749.6
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr4_+_110397661 | 0.29 |
ENSMUST00000106589.2
ENSMUST00000106587.2 ENSMUST00000106591.1 ENSMUST00000106592.1 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr1_-_158356258 | 0.26 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr4_-_155056784 | 0.26 |
ENSMUST00000131173.2
|
Plch2
|
phospholipase C, eta 2 |
| chr10_-_53647080 | 0.26 |
ENSMUST00000169866.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr1_-_78968079 | 0.25 |
ENSMUST00000049117.5
|
Gm5830
|
predicted pseudogene 5830 |
| chr2_-_33086366 | 0.25 |
ENSMUST00000049618.2
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr7_+_123123870 | 0.25 |
ENSMUST00000094053.5
|
Tnrc6a
|
trinucleotide repeat containing 6a |
| chr7_+_79273201 | 0.24 |
ENSMUST00000037315.6
|
Abhd2
|
abhydrolase domain containing 2 |
| chr5_+_138187485 | 0.24 |
ENSMUST00000110934.2
|
Cnpy4
|
canopy 4 homolog (zebrafish) |
| chr2_-_7396192 | 0.24 |
ENSMUST00000137733.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr3_-_33844255 | 0.23 |
ENSMUST00000029222.5
|
Ccdc39
|
coiled-coil domain containing 39 |
| chr2_-_79456750 | 0.21 |
ENSMUST00000041099.4
|
Neurod1
|
neurogenic differentiation 1 |
| chr1_+_53061637 | 0.21 |
ENSMUST00000027269.5
|
Mstn
|
myostatin |
| chr17_-_45733843 | 0.20 |
ENSMUST00000178179.1
|
1600014C23Rik
|
RIKEN cDNA 1600014C23 gene |
| chr8_-_3625541 | 0.19 |
ENSMUST00000136105.1
|
Pcp2
|
Purkinje cell protein 2 (L7) |
| chr12_-_24493656 | 0.19 |
ENSMUST00000073088.2
|
Gm16372
|
predicted pseudogene 16372 |
| chr1_+_161494649 | 0.19 |
ENSMUST00000086084.1
|
Tnfsf18
|
tumor necrosis factor (ligand) superfamily, member 18 |
| chr7_+_46376467 | 0.18 |
ENSMUST00000072514.1
|
Myod1
|
myogenic differentiation 1 |
| chr19_+_5088534 | 0.17 |
ENSMUST00000025811.4
|
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr15_+_101473472 | 0.17 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr17_+_37529957 | 0.17 |
ENSMUST00000097325.3
|
Olfr111
|
olfactory receptor 111 |
| chr15_+_81744848 | 0.17 |
ENSMUST00000109554.1
|
Zc3h7b
|
zinc finger CCCH type containing 7B |
| chr3_+_55782500 | 0.16 |
ENSMUST00000075422.4
|
Mab21l1
|
mab-21-like 1 (C. elegans) |
| chr4_+_154964117 | 0.15 |
ENSMUST00000030931.4
ENSMUST00000070953.4 |
Pank4
|
pantothenate kinase 4 |
| chr2_+_9882622 | 0.14 |
ENSMUST00000114919.1
|
4930412O13Rik
|
RIKEN cDNA 4930412O13 gene |
| chr7_+_64185459 | 0.13 |
ENSMUST00000177102.2
ENSMUST00000107519.1 ENSMUST00000137650.1 ENSMUST00000032737.5 ENSMUST00000107515.1 ENSMUST00000144996.1 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr6_+_77242644 | 0.13 |
ENSMUST00000159616.1
|
Lrrtm1
|
leucine rich repeat transmembrane neuronal 1 |
| chr18_+_37300799 | 0.12 |
ENSMUST00000051754.1
|
Pcdhb3
|
protocadherin beta 3 |
| chr18_+_77332394 | 0.12 |
ENSMUST00000148341.1
|
Loxhd1
|
lipoxygenase homology domains 1 |
| chr12_-_104473236 | 0.11 |
ENSMUST00000021513.4
|
Gsc
|
goosecoid homeobox |
| chr2_-_7395968 | 0.11 |
ENSMUST00000002176.6
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr12_-_11208948 | 0.10 |
ENSMUST00000049877.1
|
Msgn1
|
mesogenin 1 |
| chr18_+_69346143 | 0.09 |
ENSMUST00000114980.1
|
Tcf4
|
transcription factor 4 |
| chr11_-_102579071 | 0.09 |
ENSMUST00000107080.1
|
Gm11627
|
predicted gene 11627 |
| chr5_-_137531952 | 0.09 |
ENSMUST00000140139.1
|
Gnb2
|
guanine nucleotide binding protein (G protein), beta 2 |
| chr1_-_72874877 | 0.08 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr7_-_5014645 | 0.08 |
ENSMUST00000165320.1
|
Fiz1
|
Flt3 interacting zinc finger protein 1 |
| chr1_-_84839304 | 0.08 |
ENSMUST00000027421.6
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr18_+_24603952 | 0.07 |
ENSMUST00000025120.6
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr11_-_65269941 | 0.07 |
ENSMUST00000102635.3
|
Myocd
|
myocardin |
| chrX_-_112406779 | 0.07 |
ENSMUST00000026601.2
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr3_+_138065052 | 0.06 |
ENSMUST00000163080.2
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr13_-_100616911 | 0.04 |
ENSMUST00000168772.1
ENSMUST00000163163.1 ENSMUST00000022137.7 |
Marveld2
|
MARVEL (membrane-associating) domain containing 2 |
| chr2_-_27475622 | 0.04 |
ENSMUST00000138693.1
ENSMUST00000113941.2 ENSMUST00000077737.6 |
Brd3
|
bromodomain containing 3 |
| chr4_-_94928820 | 0.04 |
ENSMUST00000107097.2
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr2_+_71528657 | 0.03 |
ENSMUST00000126400.1
|
Dlx1
|
distal-less homeobox 1 |
| chr11_-_102579461 | 0.02 |
ENSMUST00000107081.1
|
Gm11627
|
predicted gene 11627 |
| chr6_-_129775849 | 0.01 |
ENSMUST00000095409.2
|
Gm156
|
predicted gene 156 |
| chr13_-_102905740 | 0.01 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_66468364 | 0.00 |
ENSMUST00000061620.9
|
Unc80
|
unc-80 homolog (C. elegans) |
| chr17_-_81065056 | 0.00 |
ENSMUST00000025093.4
|
Thumpd2
|
THUMP domain containing 2 |
| chr18_-_65970178 | 0.00 |
ENSMUST00000025397.5
|
Cplx4
|
complexin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Vsx2_Dlx3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0061743 | motor learning(GO:0061743) |
| 1.2 | 22.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.9 | 8.7 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.7 | 2.2 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.7 | 5.9 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.7 | 9.2 | GO:0046884 | follicle-stimulating hormone secretion(GO:0046884) |
| 0.5 | 1.6 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.5 | 4.2 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.5 | 1.6 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.4 | 5.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.4 | 1.3 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.4 | 5.4 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.3 | 4.7 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.3 | 1.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 0.9 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.3 | 2.7 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.3 | 7.7 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.2 | 3.2 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.2 | 1.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.2 | 2.6 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.9 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.2 | 0.9 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 0.8 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 1.8 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.1 | 2.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.1 | 5.5 | GO:0032094 | response to food(GO:0032094) |
| 0.1 | 3.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.1 | 0.5 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 0.5 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 1.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.3 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.6 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.1 | 1.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.8 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
| 0.1 | 0.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.4 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) cyclooxygenase pathway(GO:0019371) maintenance of blood-brain barrier(GO:0035633) |
| 0.1 | 1.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.2 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.1 | 0.7 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 1.3 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.1 | 0.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.2 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.0 | 0.6 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.5 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0060287 | regulation of cilium beat frequency(GO:0003356) determination of pancreatic left/right asymmetry(GO:0035469) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 1.1 | GO:0045109 | negative regulation of growth of symbiont involved in interaction with host(GO:0044146) intermediate filament organization(GO:0045109) |
| 0.0 | 0.8 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 1.4 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 1.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.9 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 2.4 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.7 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 1.3 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.0 | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.4 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.0 | 1.0 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.6 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0032570 | response to progesterone(GO:0032570) |
| 0.0 | 0.6 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 3.5 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.3 | 0.9 | GO:0071007 | U2-type catalytic step 2 spliceosome(GO:0071007) |
| 0.3 | 5.9 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.2 | 2.8 | GO:0071439 | clathrin complex(GO:0071439) |
| 0.2 | 2.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.2 | 0.6 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.2 | 1.6 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 22.3 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.1 | 2.6 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 1.3 | GO:0060091 | kinocilium(GO:0060091) |
| 0.1 | 1.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 1.6 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 1.7 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 3.5 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.7 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.7 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.7 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 2.9 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.5 | GO:0061702 | inflammasome complex(GO:0061702) |
| 0.0 | 1.1 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.9 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.3 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.4 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.0 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 2.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 5.3 | GO:0005635 | nuclear envelope(GO:0005635) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.4 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.7 | 5.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.4 | 4.2 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 2.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.3 | 1.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 1.6 | GO:0070728 | leucine binding(GO:0070728) |
| 0.2 | 5.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.2 | 5.9 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.2 | 1.6 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.2 | 1.8 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.2 | 0.8 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 1.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.8 | GO:0000099 | sulfur amino acid transmembrane transporter activity(GO:0000099) |
| 0.1 | 0.9 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 3.5 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 3.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 1.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 8.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 1.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.9 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.1 | 0.9 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.1 | 0.9 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.9 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 1.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 1.0 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 33.7 | GO:0008134 | transcription factor binding(GO:0008134) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 6.2 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 1.0 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 1.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 3.5 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 1.0 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.9 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.2 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 3.8 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 19.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 5.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 9.2 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.1 | 1.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.1 | 2.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 9.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 9.6 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 4.5 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.1 | 1.6 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.8 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 3.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 3.2 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |