Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
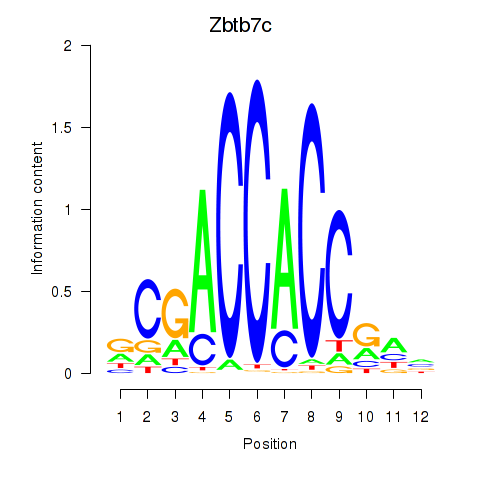
Results for Zbtb7c
Z-value: 0.95
Transcription factors associated with Zbtb7c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb7c
|
ENSMUSG00000044646.8 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb7c | mm10_v2_chr18_+_76059458_76059501 | 0.55 | 4.7e-04 | Click! |
Activity profile of Zbtb7c motif
Sorted Z-values of Zbtb7c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_100135928 | 4.73 |
ENSMUST00000107411.2
|
Krt15
|
keratin 15 |
| chr12_-_76709997 | 4.31 |
ENSMUST00000166101.1
|
Sptb
|
spectrin beta, erythrocytic |
| chr11_-_100207507 | 3.44 |
ENSMUST00000007272.7
|
Krt14
|
keratin 14 |
| chr2_+_157560078 | 3.22 |
ENSMUST00000153739.2
ENSMUST00000173595.1 ENSMUST00000109526.1 ENSMUST00000173839.1 ENSMUST00000173041.1 ENSMUST00000173793.1 ENSMUST00000172487.1 ENSMUST00000088484.5 |
Nnat
|
neuronatin |
| chr10_-_79874211 | 2.54 |
ENSMUST00000167897.1
|
BC005764
|
cDNA sequence BC005764 |
| chr14_-_54994541 | 2.54 |
ENSMUST00000153783.1
ENSMUST00000168485.1 ENSMUST00000102803.3 |
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta |
| chr14_+_118854695 | 2.49 |
ENSMUST00000100314.3
|
Cldn10
|
claudin 10 |
| chr2_-_180642681 | 2.49 |
ENSMUST00000037877.10
|
Tcfl5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr11_+_53519871 | 2.41 |
ENSMUST00000120878.2
|
Sept8
|
septin 8 |
| chr11_+_53519920 | 2.32 |
ENSMUST00000147912.1
|
Sept8
|
septin 8 |
| chr9_-_108190352 | 2.29 |
ENSMUST00000035208.7
|
Bsn
|
bassoon |
| chr11_+_80089385 | 2.28 |
ENSMUST00000108239.1
ENSMUST00000017694.5 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr4_-_108383349 | 2.18 |
ENSMUST00000053157.6
|
Fam159a
|
family with sequence similarity 159, member A |
| chr12_-_109068173 | 2.16 |
ENSMUST00000073156.7
|
Begain
|
brain-enriched guanylate kinase-associated |
| chr11_+_53519725 | 2.02 |
ENSMUST00000108987.1
ENSMUST00000121334.1 ENSMUST00000117061.1 |
Sept8
|
septin 8 |
| chr16_+_48994185 | 1.94 |
ENSMUST00000117994.1
ENSMUST00000048374.5 |
C330027C09Rik
|
RIKEN cDNA C330027C09 gene |
| chr19_-_5273080 | 1.85 |
ENSMUST00000025786.7
|
Pacs1
|
phosphofurin acidic cluster sorting protein 1 |
| chr18_-_62179948 | 1.75 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr15_+_78899755 | 1.70 |
ENSMUST00000001226.3
ENSMUST00000061239.7 ENSMUST00000109698.2 |
Sh3bp1
|
SH3-domain binding protein 1 |
| chr12_+_110279228 | 1.60 |
ENSMUST00000097228.4
|
Dio3
|
deiodinase, iodothyronine type III |
| chrX_-_142966709 | 1.60 |
ENSMUST00000041317.2
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr19_+_7417586 | 1.45 |
ENSMUST00000159348.1
|
2700081O15Rik
|
RIKEN cDNA 2700081O15 gene |
| chr7_+_45639964 | 1.42 |
ENSMUST00000148532.1
|
Mamstr
|
MEF2 activating motif and SAP domain containing transcriptional regulator |
| chr16_-_48993931 | 1.42 |
ENSMUST00000114516.1
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chrX_-_7574120 | 1.33 |
ENSMUST00000045924.7
ENSMUST00000115742.2 ENSMUST00000150787.1 |
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr15_-_82212796 | 1.29 |
ENSMUST00000179269.1
|
AI848285
|
expressed sequence AI848285 |
| chr15_-_101694299 | 1.28 |
ENSMUST00000023788.6
|
Krt6a
|
keratin 6A |
| chr17_+_47726834 | 1.24 |
ENSMUST00000024782.5
ENSMUST00000144955.1 |
Pgc
|
progastricsin (pepsinogen C) |
| chr18_-_35662180 | 1.22 |
ENSMUST00000025209.4
ENSMUST00000096573.2 |
Spata24
|
spermatogenesis associated 24 |
| chr5_+_30711564 | 1.22 |
ENSMUST00000114729.1
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr14_-_54686060 | 1.18 |
ENSMUST00000125265.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr8_+_109868586 | 1.18 |
ENSMUST00000179721.1
ENSMUST00000034175.4 |
Phlpp2
|
PH domain and leucine rich repeat protein phosphatase 2 |
| chr5_-_136883115 | 1.11 |
ENSMUST00000057497.6
ENSMUST00000111103.1 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr2_-_121271315 | 1.08 |
ENSMUST00000131245.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr3_+_107036156 | 1.08 |
ENSMUST00000052718.3
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr16_-_48994081 | 1.06 |
ENSMUST00000121869.1
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr18_+_56707725 | 1.05 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr2_-_118703963 | 1.02 |
ENSMUST00000104937.1
|
Ankrd63
|
ankyrin repeat domain 63 |
| chr5_+_30711849 | 0.98 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr15_-_85581809 | 0.87 |
ENSMUST00000023015.7
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chr8_-_122432924 | 0.86 |
ENSMUST00000017604.8
|
Cyba
|
cytochrome b-245, alpha polypeptide |
| chr1_-_86388162 | 0.83 |
ENSMUST00000027440.3
|
Nmur1
|
neuromedin U receptor 1 |
| chrX_+_36743659 | 0.83 |
ENSMUST00000047655.6
|
Slc25a43
|
solute carrier family 25, member 43 |
| chr2_-_105399286 | 0.83 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr6_+_135198034 | 0.81 |
ENSMUST00000130612.1
|
8430419L09Rik
|
RIKEN cDNA 8430419L09 gene |
| chr11_-_100441795 | 0.81 |
ENSMUST00000107398.1
|
Nt5c3b
|
5'-nucleotidase, cytosolic IIIB |
| chr15_-_101850778 | 0.81 |
ENSMUST00000023790.3
|
Krt1
|
keratin 1 |
| chr15_-_101680281 | 0.81 |
ENSMUST00000023786.5
|
Krt6b
|
keratin 6B |
| chr11_+_49794157 | 0.80 |
ENSMUST00000020629.4
|
Gfpt2
|
glutamine fructose-6-phosphate transaminase 2 |
| chr4_-_120287349 | 0.80 |
ENSMUST00000102656.3
|
Foxo6
|
forkhead box O6 |
| chr8_+_25911670 | 0.79 |
ENSMUST00000120653.1
ENSMUST00000126226.1 |
Kcnu1
|
potassium channel, subfamily U, member 1 |
| chr1_-_153332724 | 0.77 |
ENSMUST00000027752.8
|
Lamc1
|
laminin, gamma 1 |
| chr10_-_93589621 | 0.76 |
ENSMUST00000020203.6
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr11_+_68432112 | 0.75 |
ENSMUST00000021283.7
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5, p101 |
| chr13_-_55513427 | 0.74 |
ENSMUST00000069929.6
ENSMUST00000069968.6 ENSMUST00000131306.1 ENSMUST00000046246.6 |
Pdlim7
|
PDZ and LIM domain 7 |
| chr12_-_110840905 | 0.73 |
ENSMUST00000177224.1
ENSMUST00000084974.4 ENSMUST00000070565.8 |
Stk30
|
serine/threonine kinase 30 |
| chr5_+_122158265 | 0.72 |
ENSMUST00000102528.4
ENSMUST00000086294.6 |
Ppp1cc
|
protein phosphatase 1, catalytic subunit, gamma isoform |
| chr1_-_21961581 | 0.72 |
ENSMUST00000029667.6
ENSMUST00000173058.1 ENSMUST00000173404.1 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr1_-_45890078 | 0.71 |
ENSMUST00000183590.1
|
Gm5269
|
predicted gene 5269 |
| chr12_+_31265234 | 0.71 |
ENSMUST00000169088.1
|
Lamb1
|
laminin B1 |
| chr16_+_11984581 | 0.71 |
ENSMUST00000170672.2
ENSMUST00000023138.7 |
Shisa9
|
shisa homolog 9 (Xenopus laevis) |
| chr17_+_34894515 | 0.67 |
ENSMUST00000052778.8
|
Zbtb12
|
zinc finger and BTB domain containing 12 |
| chr9_-_85327110 | 0.67 |
ENSMUST00000034802.8
|
Fam46a
|
family with sequence similarity 46, member A |
| chr14_+_25607797 | 0.65 |
ENSMUST00000160229.1
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr5_-_148928619 | 0.62 |
ENSMUST00000149169.1
ENSMUST00000047257.8 |
Katnal1
|
katanin p60 subunit A-like 1 |
| chr8_+_72240052 | 0.61 |
ENSMUST00000145213.1
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr11_+_101176041 | 0.60 |
ENSMUST00000103109.3
|
Cntnap1
|
contactin associated protein-like 1 |
| chr2_+_158794807 | 0.60 |
ENSMUST00000029186.7
ENSMUST00000109478.2 ENSMUST00000156893.1 |
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr11_+_115564434 | 0.60 |
ENSMUST00000021085.4
|
Nup85
|
nucleoporin 85 |
| chr12_+_31265279 | 0.59 |
ENSMUST00000002979.8
ENSMUST00000170495.1 |
Lamb1
|
laminin B1 |
| chr8_+_72240315 | 0.58 |
ENSMUST00000126885.1
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr3_+_134236483 | 0.58 |
ENSMUST00000181904.1
ENSMUST00000053048.9 |
Cxxc4
|
CXXC finger 4 |
| chr19_-_41848076 | 0.57 |
ENSMUST00000059231.2
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr14_-_54686605 | 0.57 |
ENSMUST00000147714.1
|
Acin1
|
apoptotic chromatin condensation inducer 1 |
| chr11_+_70000578 | 0.57 |
ENSMUST00000019362.8
|
Dvl2
|
dishevelled 2, dsh homolog (Drosophila) |
| chr8_+_72240018 | 0.56 |
ENSMUST00000003117.8
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr19_-_53371766 | 0.56 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr7_+_5056856 | 0.56 |
ENSMUST00000131368.1
ENSMUST00000123956.1 |
Ccdc106
|
coiled-coil domain containing 106 |
| chr5_-_134614953 | 0.55 |
ENSMUST00000036362.6
ENSMUST00000077636.4 |
Lat2
|
linker for activation of T cells family, member 2 |
| chr7_-_6331235 | 0.55 |
ENSMUST00000127658.1
ENSMUST00000062765.7 |
Zfp583
|
zinc finger protein 583 |
| chr2_-_121271403 | 0.52 |
ENSMUST00000110648.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr1_-_165460430 | 0.51 |
ENSMUST00000027856.7
|
Dcaf6
|
DDB1 and CUL4 associated factor 6 |
| chr11_-_115514374 | 0.49 |
ENSMUST00000021083.6
|
Hn1
|
hematological and neurological expressed sequence 1 |
| chr7_+_5056706 | 0.48 |
ENSMUST00000144802.1
|
Ccdc106
|
coiled-coil domain containing 106 |
| chr18_+_86394952 | 0.47 |
ENSMUST00000058829.2
|
Neto1
|
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_+_55406163 | 0.46 |
ENSMUST00000042986.8
|
Plcl1
|
phospholipase C-like 1 |
| chr2_-_121271341 | 0.44 |
ENSMUST00000110647.1
|
Trp53bp1
|
transformation related protein 53 binding protein 1 |
| chr4_+_8691303 | 0.41 |
ENSMUST00000051558.3
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr14_-_67008834 | 0.40 |
ENSMUST00000111115.1
ENSMUST00000022634.8 |
Bnip3l
|
BCL2/adenovirus E1B interacting protein 3-like |
| chr7_-_19310035 | 0.40 |
ENSMUST00000003640.2
|
Fosb
|
FBJ osteosarcoma oncogene B |
| chr18_+_34777008 | 0.39 |
ENSMUST00000043775.7
|
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B |
| chr8_-_34965631 | 0.38 |
ENSMUST00000033929.4
|
Tnks
|
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr19_+_40612392 | 0.38 |
ENSMUST00000134063.1
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr6_+_135197977 | 0.37 |
ENSMUST00000111915.1
ENSMUST00000111916.1 |
8430419L09Rik
|
RIKEN cDNA 8430419L09 gene |
| chr11_+_75532127 | 0.36 |
ENSMUST00000127226.1
|
Slc43a2
|
solute carrier family 43, member 2 |
| chr2_-_45117349 | 0.35 |
ENSMUST00000176438.2
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_85035487 | 0.35 |
ENSMUST00000028465.7
|
P2rx3
|
purinergic receptor P2X, ligand-gated ion channel, 3 |
| chr7_-_19715395 | 0.35 |
ENSMUST00000032555.9
ENSMUST00000093552.5 |
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr2_-_13793793 | 0.34 |
ENSMUST00000003509.8
|
St8sia6
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
| chr2_-_32312162 | 0.34 |
ENSMUST00000155269.1
|
Dnm1
|
dynamin 1 |
| chrX_+_20617503 | 0.33 |
ENSMUST00000115375.1
ENSMUST00000115374.1 ENSMUST00000084383.3 |
Rbm10
|
RNA binding motif protein 10 |
| chr9_+_67840386 | 0.32 |
ENSMUST00000077879.5
|
Vps13c
|
vacuolar protein sorting 13C (yeast) |
| chr2_+_121955964 | 0.31 |
ENSMUST00000036647.6
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_-_102447647 | 0.30 |
ENSMUST00000049057.4
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chrX_-_41911877 | 0.28 |
ENSMUST00000047037.8
|
Thoc2
|
THO complex 2 |
| chr9_+_66350465 | 0.28 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr12_+_110841021 | 0.25 |
ENSMUST00000181348.1
|
4921507G05Rik
|
RIKEN cDNA 4921507G05 gene |
| chr18_-_61400363 | 0.24 |
ENSMUST00000063307.5
ENSMUST00000075299.6 |
Ppargc1b
|
peroxisome proliferative activated receptor, gamma, coactivator 1 beta |
| chr7_-_13038201 | 0.24 |
ENSMUST00000005714.7
ENSMUST00000156389.1 ENSMUST00000165394.2 |
Ube2m
|
ubiquitin-conjugating enzyme E2M |
| chr1_+_136018062 | 0.23 |
ENSMUST00000117950.1
|
Tmem9
|
transmembrane protein 9 |
| chr8_+_33386325 | 0.20 |
ENSMUST00000078058.3
ENSMUST00000070340.4 |
Purg
|
purine-rich element binding protein G |
| chr10_+_111164794 | 0.20 |
ENSMUST00000105275.1
ENSMUST00000095310.1 |
Osbpl8
|
oxysterol binding protein-like 8 |
| chr11_+_75531690 | 0.19 |
ENSMUST00000149727.1
ENSMUST00000042561.7 ENSMUST00000108433.1 ENSMUST00000143035.1 |
Slc43a2
|
solute carrier family 43, member 2 |
| chr10_-_79766872 | 0.18 |
ENSMUST00000047203.8
|
Rnf126
|
ring finger protein 126 |
| chr16_+_17561885 | 0.18 |
ENSMUST00000171002.1
ENSMUST00000023441.4 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr18_-_5334364 | 0.15 |
ENSMUST00000063989.5
|
Zfp438
|
zinc finger protein 438 |
| chr11_+_85832551 | 0.14 |
ENSMUST00000000095.6
|
Tbx2
|
T-box 2 |
| chrX_-_72274747 | 0.14 |
ENSMUST00000064780.3
|
Gabre
|
gamma-aminobutyric acid (GABA) A receptor, subunit epsilon |
| chr5_+_108268897 | 0.13 |
ENSMUST00000031190.4
|
Dr1
|
down-regulator of transcription 1 |
| chr2_-_72980402 | 0.12 |
ENSMUST00000066003.6
ENSMUST00000102689.3 |
Sp3
|
trans-acting transcription factor 3 |
| chrX_+_152233228 | 0.11 |
ENSMUST00000112588.2
ENSMUST00000082177.6 |
Kdm5c
|
lysine (K)-specific demethylase 5C |
| chr12_+_29938036 | 0.11 |
ENSMUST00000122328.1
ENSMUST00000118321.1 |
Pxdn
|
peroxidasin homolog (Drosophila) |
| chr4_-_128962420 | 0.11 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr3_+_87906321 | 0.10 |
ENSMUST00000005017.8
|
Hdgf
|
hepatoma-derived growth factor |
| chr4_-_116821501 | 0.10 |
ENSMUST00000055436.3
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr2_-_119477613 | 0.07 |
ENSMUST00000110808.1
ENSMUST00000049920.7 |
Ino80
|
INO80 homolog (S. cerevisiae) |
| chr12_-_76369586 | 0.07 |
ENSMUST00000176278.1
|
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr11_-_71004387 | 0.06 |
ENSMUST00000124464.1
ENSMUST00000108527.1 |
Dhx33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr8_+_95825353 | 0.05 |
ENSMUST00000074053.4
|
Gm10094
|
predicted gene 10094 |
| chr12_-_76369385 | 0.04 |
ENSMUST00000176187.1
ENSMUST00000167011.1 ENSMUST00000176967.1 |
Zbtb25
|
zinc finger and BTB domain containing 25 |
| chr5_-_137786651 | 0.03 |
ENSMUST00000031740.9
|
Mepce
|
methylphosphate capping enzyme |
| chr10_+_93589413 | 0.02 |
ENSMUST00000181835.1
|
4933408J17Rik
|
RIKEN cDNA 4933408J17 gene |
| chr5_-_137786681 | 0.02 |
ENSMUST00000132726.1
|
Mepce
|
methylphosphate capping enzyme |
| chr3_-_96727453 | 0.01 |
ENSMUST00000141377.1
ENSMUST00000125183.1 |
Polr3c
|
polymerase (RNA) III (DNA directed) polypeptide C |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb7c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.5 | 2.5 | GO:0031444 | regulation of twitch skeletal muscle contraction(GO:0014724) slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.5 | 3.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.4 | 1.8 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.4 | 1.1 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.3 | 1.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 0.9 | GO:0002019 | regulation of renal output by angiotensin(GO:0002019) |
| 0.2 | 1.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.2 | 1.8 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.2 | 0.9 | GO:0072054 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) renal outer medulla development(GO:0072054) inner medullary collecting duct development(GO:0072061) |
| 0.2 | 1.9 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.2 | 1.6 | GO:0019336 | phenol-containing compound catabolic process(GO:0019336) |
| 0.2 | 0.5 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 2.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.1 | 0.6 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.1 | 2.0 | GO:0045830 | positive regulation of isotype switching(GO:0045830) negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.1 | 0.4 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.1 | 1.7 | GO:0043485 | endosome to melanosome transport(GO:0035646) endosome to pigment granule transport(GO:0043485) pigment granule maturation(GO:0048757) |
| 0.1 | 0.6 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 0.4 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.1 | 1.3 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 2.1 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.1 | 4.3 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.1 | 0.6 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.8 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.6 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.1 | 0.4 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.1 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.4 | GO:0072718 | response to cisplatin(GO:0072718) |
| 0.1 | 0.8 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.3 | GO:1904923 | regulation of mitophagy in response to mitochondrial depolarization(GO:1904923) |
| 0.1 | 0.4 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.3 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.2 | GO:0010694 | positive regulation of alkaline phosphatase activity(GO:0010694) |
| 0.0 | 0.1 | GO:0007521 | muscle cell fate determination(GO:0007521) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.8 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 1.4 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.7 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.3 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 3.2 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 2.5 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.7 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 1.9 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.8 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.2 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.6 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.3 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 2.4 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.6 | 4.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.5 | 2.1 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.3 | 2.5 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.2 | 1.8 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.2 | 0.8 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.2 | 5.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.2 | 0.8 | GO:0034715 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 2.5 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.1 | 2.5 | GO:0032982 | muscle myosin complex(GO:0005859) myosin filament(GO:0032982) |
| 0.1 | 1.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.7 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.9 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 0.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 3.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.0 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 1.7 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 1.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.5 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 3.2 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.3 | GO:0000445 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 7.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 3.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.5 | 1.6 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.4 | 1.8 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) bradykinin receptor binding(GO:0031711) |
| 0.3 | 2.0 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.3 | 1.3 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 0.8 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.2 | 0.8 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 2.5 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 2.5 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 4.3 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 2.4 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.1 | 0.8 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.6 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.1 | 0.9 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 1.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 4.3 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 1.2 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.8 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.4 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 1.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.3 | GO:0031749 | D2 dopamine receptor binding(GO:0031749) |
| 0.0 | 0.5 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.8 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.8 | GO:0008188 | neuropeptide receptor activity(GO:0008188) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.4 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 1.2 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 5.8 | GO:0005525 | GTP binding(GO:0005525) |
| 0.0 | 2.2 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.6 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.7 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 1.9 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 1.3 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 3.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.7 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.9 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.6 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 4.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 1.6 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.1 | 0.8 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 3.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.9 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.6 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |