Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
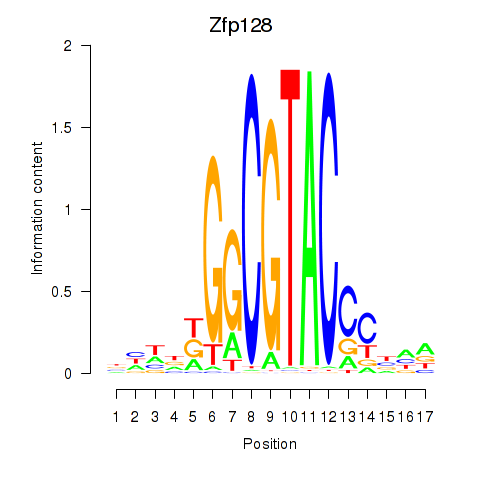
Results for Zfp128
Z-value: 0.87
Transcription factors associated with Zfp128
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp128
|
ENSMUSG00000060397.6 | zinc finger protein 128 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp128 | mm10_v2_chr7_+_12881165_12881204 | 0.54 | 7.0e-04 | Click! |
Activity profile of Zfp128 motif
Sorted Z-values of Zfp128 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_76669811 | 4.05 |
ENSMUST00000037824.4
|
Foxh1
|
forkhead box H1 |
| chr7_-_45526146 | 3.71 |
ENSMUST00000167273.1
ENSMUST00000042105.8 |
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A |
| chr15_-_96642883 | 3.61 |
ENSMUST00000088452.4
|
Slc38a1
|
solute carrier family 38, member 1 |
| chr11_+_32300069 | 3.21 |
ENSMUST00000020535.1
|
Hbq1a
|
hemoglobin, theta 1A |
| chr4_+_126556935 | 2.50 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr6_+_128362919 | 2.43 |
ENSMUST00000073316.6
|
Foxm1
|
forkhead box M1 |
| chr4_+_126556994 | 2.39 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr5_-_138170992 | 2.29 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr5_+_137761680 | 2.26 |
ENSMUST00000110983.2
ENSMUST00000031738.4 |
Tsc22d4
|
TSC22 domain family, member 4 |
| chrX_-_134583114 | 2.21 |
ENSMUST00000113213.1
ENSMUST00000033617.6 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr5_-_138171248 | 2.14 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr11_+_9191934 | 2.08 |
ENSMUST00000042740.6
|
Abca13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr5_-_138171216 | 1.96 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr6_-_128362812 | 1.24 |
ENSMUST00000112152.1
ENSMUST00000057421.8 ENSMUST00000112151.1 |
Rhno1
|
RAD9-HUS1-RAD1 interacting nuclear orphan 1 |
| chr7_+_45434876 | 1.22 |
ENSMUST00000107766.1
|
Gys1
|
glycogen synthase 1, muscle |
| chr6_-_118562226 | 1.17 |
ENSMUST00000112830.1
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr9_+_3335470 | 1.10 |
ENSMUST00000053407.5
|
Alkbh8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr13_-_59769751 | 1.06 |
ENSMUST00000057115.6
|
Isca1
|
iron-sulfur cluster assembly 1 homolog (S. cerevisiae) |
| chr18_-_37644185 | 0.97 |
ENSMUST00000066272.4
|
Taf7
|
TAF7 RNA polymerase II, TATA box binding protein (TBP)-associated factor |
| chr15_+_76879232 | 0.88 |
ENSMUST00000023179.5
|
Zfp7
|
zinc finger protein 7 |
| chr3_+_103576081 | 0.82 |
ENSMUST00000183637.1
ENSMUST00000117221.2 ENSMUST00000118117.1 ENSMUST00000118563.2 |
Syt6
|
synaptotagmin VI |
| chr8_+_83715177 | 0.77 |
ENSMUST00000019576.8
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr5_-_73632421 | 0.70 |
ENSMUST00000087177.2
|
Lrrc66
|
leucine rich repeat containing 66 |
| chr5_+_144545883 | 0.70 |
ENSMUST00000071782.6
|
Nptx2
|
neuronal pentraxin 2 |
| chr2_+_5137756 | 0.70 |
ENSMUST00000027988.7
|
Ccdc3
|
coiled-coil domain containing 3 |
| chr8_+_83715504 | 0.68 |
ENSMUST00000109810.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr7_-_45434590 | 0.62 |
ENSMUST00000107771.3
ENSMUST00000141761.1 |
Ruvbl2
|
RuvB-like protein 2 |
| chr1_-_195092242 | 0.58 |
ENSMUST00000162650.1
ENSMUST00000160817.1 ENSMUST00000162614.1 ENSMUST00000016637.6 |
Cd46
|
CD46 antigen, complement regulatory protein |
| chr8_-_126945841 | 0.53 |
ENSMUST00000179857.1
|
Tomm20
|
translocase of outer mitochondrial membrane 20 homolog (yeast) |
| chr14_-_105896819 | 0.53 |
ENSMUST00000022709.4
|
Spry2
|
sprouty homolog 2 (Drosophila) |
| chr8_+_83715239 | 0.51 |
ENSMUST00000172396.1
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 |
| chr19_-_47536997 | 0.50 |
ENSMUST00000182808.1
ENSMUST00000049369.9 |
Obfc1
|
oligonucleotide/oligosaccharide-binding fold containing 1 |
| chr3_+_79568192 | 0.46 |
ENSMUST00000091023.1
|
4930589L23Rik
|
RIKEN cDNA 4930589L23 gene |
| chr3_+_37420273 | 0.46 |
ENSMUST00000029277.8
|
Spata5
|
spermatogenesis associated 5 |
| chr9_+_21411824 | 0.46 |
ENSMUST00000002902.6
|
Qtrt1
|
queuine tRNA-ribosyltransferase 1 |
| chr6_-_145210791 | 0.44 |
ENSMUST00000111728.1
ENSMUST00000060797.7 |
Casc1
|
cancer susceptibility candidate 1 |
| chr7_-_44929410 | 0.41 |
ENSMUST00000107857.3
ENSMUST00000085399.6 ENSMUST00000167930.1 ENSMUST00000166972.1 |
Ap2a1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr14_+_54894133 | 0.36 |
ENSMUST00000116476.2
ENSMUST00000022808.7 ENSMUST00000150975.1 |
Pabpn1
|
poly(A) binding protein, nuclear 1 |
| chr7_+_45526330 | 0.36 |
ENSMUST00000120985.1
ENSMUST00000051810.8 |
Plekha4
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
| chr2_-_127143306 | 0.35 |
ENSMUST00000110386.1
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
| chr7_-_79386943 | 0.34 |
ENSMUST00000053718.8
ENSMUST00000179243.1 |
Rlbp1
|
retinaldehyde binding protein 1 |
| chr9_-_55048544 | 0.31 |
ENSMUST00000034854.6
|
Chrnb4
|
cholinergic receptor, nicotinic, beta polypeptide 4 |
| chr7_+_24112314 | 0.30 |
ENSMUST00000120006.1
ENSMUST00000005413.3 |
Zfp112
|
zinc finger protein 112 |
| chr2_+_71055731 | 0.30 |
ENSMUST00000154704.1
ENSMUST00000135357.1 ENSMUST00000064141.5 ENSMUST00000112159.2 ENSMUST00000102701.3 |
Dcaf17
|
DDB1 and CUL4 associated factor 17 |
| chr2_-_127143410 | 0.28 |
ENSMUST00000132773.1
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1 |
| chr4_+_15881255 | 0.25 |
ENSMUST00000029876.1
|
Calb1
|
calbindin 1 |
| chr15_+_76246747 | 0.21 |
ENSMUST00000023225.6
|
Grina
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_-_35176039 | 0.20 |
ENSMUST00000119847.1
ENSMUST00000034539.5 |
Dcps
|
decapping enzyme, scavenger |
| chrX_-_53776392 | 0.17 |
ENSMUST00000165081.1
ENSMUST00000071711.3 |
Zfp36l3
|
zinc finger protein 36, C3H type-like 3 |
| chr17_+_33843085 | 0.13 |
ENSMUST00000002379.8
|
Cd320
|
CD320 antigen |
| chr3_-_79567679 | 0.08 |
ENSMUST00000076136.4
|
Fnip2
|
folliculin interacting protein 2 |
| chr3_-_79567771 | 0.06 |
ENSMUST00000133154.1
|
Fnip2
|
folliculin interacting protein 2 |
| chr7_-_21427941 | 0.02 |
ENSMUST00000177741.1
|
Gm6882
|
predicted gene 6882 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp128
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 1.0 | 4.0 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.7 | 2.2 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.6 | 6.4 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 3.6 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.4 | 4.9 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.3 | 0.6 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.2 | 2.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.2 | 2.4 | GO:2000781 | positive regulation of double-strand break repair(GO:2000781) |
| 0.2 | 0.6 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.4 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.8 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.1 | 0.4 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.1 | 0.5 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 1.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 0.5 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 0.2 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.7 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.0 | 2.0 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 1.0 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.2 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 1.2 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 1.1 | GO:0071479 | cellular response to ionizing radiation(GO:0071479) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.0 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.4 | 6.4 | GO:0042555 | MCM complex(GO:0042555) |
| 0.3 | 3.7 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.1 | 0.6 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 1.0 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.1 | 0.6 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 0.5 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.8 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 1.2 | GO:0016234 | inclusion body(GO:0016234) |
| 0.0 | 2.0 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.9 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.7 | 3.6 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.4 | 1.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.2 | 1.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.2 | 1.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 0.5 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.2 | 7.0 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 4.0 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 3.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.1 | 0.5 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 2.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 1.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 2.0 | GO:0008026 | ATP-dependent helicase activity(GO:0008026) purine NTP-dependent helicase activity(GO:0070035) |
| 0.0 | 0.3 | GO:0019841 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.4 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.3 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 2.1 | GO:0042626 | ATPase activity, coupled to transmembrane movement of substances(GO:0042626) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 11.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 3.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 2.2 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 4.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.4 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 4.0 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 3.7 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.1 | 3.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 2.2 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.6 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.6 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |