Project
avrg: GSE58827: Dynamics of the Mouse Liver
Navigation
Downloads
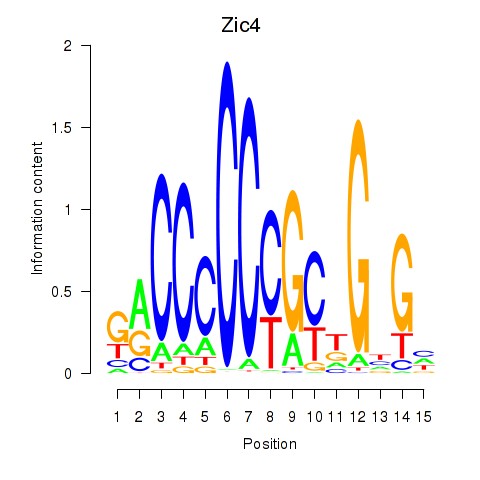
Results for Zic4
Z-value: 0.53
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic4
|
ENSMUSG00000036972.8 | zinc finger protein of the cerebellum 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | mm10_v2_chr9_+_91378636_91378646 | -0.02 | 9.2e-01 | Click! |
Activity profile of Zic4 motif
Sorted Z-values of Zic4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_107986360 | 1.83 |
ENSMUST00000066530.6
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr3_-_107986408 | 1.82 |
ENSMUST00000012348.2
|
Gstm2
|
glutathione S-transferase, mu 2 |
| chr14_-_64455903 | 1.56 |
ENSMUST00000067927.7
|
Msra
|
methionine sulfoxide reductase A |
| chr11_+_99041237 | 1.36 |
ENSMUST00000017637.6
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr4_+_141368116 | 1.24 |
ENSMUST00000006380.4
|
Fam131c
|
family with sequence similarity 131, member C |
| chr12_-_102704896 | 1.20 |
ENSMUST00000178697.1
ENSMUST00000046518.5 |
Itpk1
|
inositol 1,3,4-triphosphate 5/6 kinase |
| chr4_-_57143437 | 1.20 |
ENSMUST00000095076.3
ENSMUST00000030142.3 |
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b |
| chr11_+_77515104 | 1.12 |
ENSMUST00000094004.4
|
Abhd15
|
abhydrolase domain containing 15 |
| chr17_-_57059795 | 1.09 |
ENSMUST00000040280.7
|
Slc25a23
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 23 |
| chr11_-_78422217 | 1.05 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr1_-_51915968 | 0.95 |
ENSMUST00000046390.7
|
Myo1b
|
myosin IB |
| chr6_-_59426279 | 0.94 |
ENSMUST00000051065.4
|
Gprin3
|
GPRIN family member 3 |
| chr11_-_87987528 | 0.93 |
ENSMUST00000020775.2
|
Dynll2
|
dynein light chain LC8-type 2 |
| chr14_-_118052235 | 0.92 |
ENSMUST00000022725.2
|
Dct
|
dopachrome tautomerase |
| chr14_-_47189406 | 0.89 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr14_+_33941021 | 0.82 |
ENSMUST00000100720.1
|
Gdf2
|
growth differentiation factor 2 |
| chr8_-_64849818 | 0.80 |
ENSMUST00000034017.7
|
Klhl2
|
kelch-like 2, Mayven |
| chr11_-_101171302 | 0.80 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr2_+_32535724 | 0.78 |
ENSMUST00000133366.1
|
Fam102a
|
family with sequence similarity 102, member A |
| chr9_+_58554799 | 0.77 |
ENSMUST00000098676.2
|
Gm10657
|
predicted gene 10657 |
| chr17_-_45593626 | 0.76 |
ENSMUST00000163905.1
ENSMUST00000167692.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr8_-_45382198 | 0.74 |
ENSMUST00000093526.6
|
Fam149a
|
family with sequence similarity 149, member A |
| chr9_+_46012810 | 0.73 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr16_+_20733104 | 0.73 |
ENSMUST00000115423.1
ENSMUST00000007171.6 |
Chrd
|
chordin |
| chr17_-_34031644 | 0.72 |
ENSMUST00000171872.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr11_+_57801575 | 0.71 |
ENSMUST00000020826.5
|
Sap30l
|
SAP30-like |
| chr7_+_143107252 | 0.70 |
ENSMUST00000009689.4
|
Kcnq1
|
potassium voltage-gated channel, subfamily Q, member 1 |
| chr7_+_127800604 | 0.70 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_-_152951456 | 0.69 |
ENSMUST00000123121.2
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr4_-_72200833 | 0.69 |
ENSMUST00000102848.2
ENSMUST00000072695.6 ENSMUST00000107337.1 ENSMUST00000074216.7 |
Tle1
|
transducin-like enhancer of split 1, homolog of Drosophila E(spl) |
| chr15_-_39857459 | 0.67 |
ENSMUST00000022915.3
ENSMUST00000110306.1 |
Dpys
|
dihydropyrimidinase |
| chr17_-_34031544 | 0.66 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr17_-_34031684 | 0.64 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr15_+_7129557 | 0.63 |
ENSMUST00000067190.5
ENSMUST00000164529.1 |
Lifr
|
leukemia inhibitory factor receptor |
| chr7_+_127800844 | 0.63 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_+_139543889 | 0.62 |
ENSMUST00000174792.1
ENSMUST00000031523.8 |
Uncx
|
UNC homeobox |
| chr7_+_27591705 | 0.61 |
ENSMUST00000167435.1
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr19_+_4855129 | 0.61 |
ENSMUST00000119694.1
|
Ctsf
|
cathepsin F |
| chr2_+_156065738 | 0.59 |
ENSMUST00000137966.1
|
Spag4
|
sperm associated antigen 4 |
| chr7_-_34654342 | 0.58 |
ENSMUST00000108069.1
|
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr17_-_45595842 | 0.58 |
ENSMUST00000164618.1
ENSMUST00000097317.3 ENSMUST00000170113.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr9_+_46012822 | 0.57 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr17_+_26113286 | 0.57 |
ENSMUST00000025010.7
|
Tmem8
|
transmembrane protein 8 (five membrane-spanning domains) |
| chr4_+_148130883 | 0.56 |
ENSMUST00000084129.2
|
Mad2l2
|
MAD2 mitotic arrest deficient-like 2 |
| chr9_-_58555129 | 0.56 |
ENSMUST00000165365.1
|
Cd276
|
CD276 antigen |
| chr7_-_137314394 | 0.56 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr2_+_147187424 | 0.54 |
ENSMUST00000144411.1
|
6430503K07Rik
|
RIKEN cDNA 6430503K07 gene |
| chr11_+_101468164 | 0.52 |
ENSMUST00000001347.6
|
Rnd2
|
Rho family GTPase 2 |
| chr12_-_57546121 | 0.52 |
ENSMUST00000044380.6
|
Foxa1
|
forkhead box A1 |
| chr5_-_66150898 | 0.50 |
ENSMUST00000113725.1
ENSMUST00000094757.2 |
Rbm47
|
RNA binding motif protein 47 |
| chr14_-_29721835 | 0.50 |
ENSMUST00000022567.7
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3 |
| chr3_-_95015416 | 0.50 |
ENSMUST00000132195.1
|
Zfp687
|
zinc finger protein 687 |
| chr7_+_28766747 | 0.50 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr2_+_120977017 | 0.49 |
ENSMUST00000067582.7
|
Tmem62
|
transmembrane protein 62 |
| chr2_-_130642770 | 0.47 |
ENSMUST00000045761.6
|
Lzts3
|
leucine zipper, putative tumor suppressor family member 3 |
| chr2_+_32535315 | 0.47 |
ENSMUST00000133512.1
ENSMUST00000048375.5 |
Fam102a
|
family with sequence similarity 102, member A |
| chr2_-_152951688 | 0.47 |
ENSMUST00000109811.3
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr18_-_77565050 | 0.46 |
ENSMUST00000182153.1
ENSMUST00000182146.1 ENSMUST00000026494.7 ENSMUST00000182024.1 |
Rnf165
|
ring finger protein 165 |
| chr9_-_29412204 | 0.46 |
ENSMUST00000115237.1
|
Ntm
|
neurotrimin |
| chr7_+_27591513 | 0.45 |
ENSMUST00000108344.2
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr2_+_156065180 | 0.45 |
ENSMUST00000038860.5
|
Spag4
|
sperm associated antigen 4 |
| chr2_-_152951547 | 0.45 |
ENSMUST00000037715.6
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr2_-_29253001 | 0.45 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr5_+_24426831 | 0.45 |
ENSMUST00000155598.1
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr2_-_167062607 | 0.45 |
ENSMUST00000128676.1
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr18_+_12333953 | 0.44 |
ENSMUST00000092070.6
|
Lama3
|
laminin, alpha 3 |
| chr10_+_106470281 | 0.43 |
ENSMUST00000029404.9
ENSMUST00000169303.1 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr17_-_85688252 | 0.43 |
ENSMUST00000024947.7
ENSMUST00000163568.2 |
Six2
|
sine oculis-related homeobox 2 |
| chr17_-_45595502 | 0.42 |
ENSMUST00000171081.1
ENSMUST00000172301.1 ENSMUST00000167332.1 ENSMUST00000170488.1 ENSMUST00000167195.1 ENSMUST00000064889.6 ENSMUST00000051574.6 ENSMUST00000164217.1 |
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr19_-_4477447 | 0.42 |
ENSMUST00000059295.3
|
Syt12
|
synaptotagmin XII |
| chr15_-_33687840 | 0.41 |
ENSMUST00000042021.3
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5 |
| chr4_-_108217897 | 0.41 |
ENSMUST00000106690.1
ENSMUST00000043793.6 |
Zyg11a
|
zyg-11 family member A, cell cycle regulator |
| chr5_-_113310697 | 0.40 |
ENSMUST00000154248.1
|
Sgsm1
|
small G protein signaling modulator 1 |
| chr7_-_19822698 | 0.39 |
ENSMUST00000120537.1
|
Bcl3
|
B cell leukemia/lymphoma 3 |
| chr2_-_144331695 | 0.39 |
ENSMUST00000103171.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr7_-_97579382 | 0.39 |
ENSMUST00000151840.1
ENSMUST00000135998.1 ENSMUST00000144858.1 ENSMUST00000146605.1 ENSMUST00000072725.5 ENSMUST00000138060.1 ENSMUST00000154853.1 ENSMUST00000136757.1 ENSMUST00000124552.1 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr17_-_26508463 | 0.38 |
ENSMUST00000025025.6
|
Dusp1
|
dual specificity phosphatase 1 |
| chr9_-_107710475 | 0.37 |
ENSMUST00000080560.3
|
Sema3f
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr13_+_34037619 | 0.35 |
ENSMUST00000040656.6
|
Bphl
|
biphenyl hydrolase-like (serine hydrolase, breast epithelial mucin-associated antigen) |
| chr10_+_127380591 | 0.34 |
ENSMUST00000166820.1
|
R3hdm2
|
R3H domain containing 2 |
| chr11_-_55607733 | 0.34 |
ENSMUST00000108853.1
ENSMUST00000075603.4 |
Glra1
|
glycine receptor, alpha 1 subunit |
| chr10_+_127380799 | 0.33 |
ENSMUST00000111628.2
|
R3hdm2
|
R3H domain containing 2 |
| chr10_+_127739516 | 0.32 |
ENSMUST00000054287.7
|
Zbtb39
|
zinc finger and BTB domain containing 39 |
| chr7_-_63938862 | 0.32 |
ENSMUST00000063694.8
|
Klf13
|
Kruppel-like factor 13 |
| chr1_+_75546449 | 0.32 |
ENSMUST00000150142.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr19_-_6235804 | 0.32 |
ENSMUST00000025695.9
|
Ppp2r5b
|
protein phosphatase 2, regulatory subunit B (B56), beta isoform |
| chr5_-_113310729 | 0.32 |
ENSMUST00000112325.1
ENSMUST00000048112.6 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr4_-_156197479 | 0.31 |
ENSMUST00000075787.6
ENSMUST00000180572.1 |
Agrn
|
agrin |
| chr13_-_95223045 | 0.31 |
ENSMUST00000162292.1
|
Pde8b
|
phosphodiesterase 8B |
| chr4_+_62360695 | 0.30 |
ENSMUST00000084526.5
|
Slc31a1
|
solute carrier family 31, member 1 |
| chr2_-_167062981 | 0.30 |
ENSMUST00000048988.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr11_+_97663366 | 0.30 |
ENSMUST00000044730.5
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6 |
| chr2_+_164879358 | 0.29 |
ENSMUST00000041643.3
|
Pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr6_-_115251839 | 0.29 |
ENSMUST00000032462.6
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr6_+_65671590 | 0.28 |
ENSMUST00000054351.4
|
Ndnf
|
neuron-derived neurotrophic factor |
| chr6_+_141524379 | 0.28 |
ENSMUST00000032362.9
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr13_-_48625571 | 0.28 |
ENSMUST00000035824.9
|
Ptpdc1
|
protein tyrosine phosphatase domain containing 1 |
| chr4_+_152338887 | 0.27 |
ENSMUST00000005175.4
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr5_+_43233463 | 0.27 |
ENSMUST00000169035.1
ENSMUST00000166713.1 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr6_-_39118211 | 0.27 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr4_+_129513581 | 0.27 |
ENSMUST00000062356.6
|
Marcksl1
|
MARCKS-like 1 |
| chr2_+_119325784 | 0.26 |
ENSMUST00000102517.3
|
Dll4
|
delta-like 4 (Drosophila) |
| chr2_-_25234227 | 0.26 |
ENSMUST00000006638.7
|
Slc34a3
|
solute carrier family 34 (sodium phosphate), member 3 |
| chr4_-_151996113 | 0.26 |
ENSMUST00000055688.9
|
Phf13
|
PHD finger protein 13 |
| chr1_-_168431695 | 0.26 |
ENSMUST00000176790.1
|
Pbx1
|
pre B cell leukemia homeobox 1 |
| chrX_-_155623325 | 0.25 |
ENSMUST00000038665.5
|
Ptchd1
|
patched domain containing 1 |
| chr8_-_36613937 | 0.25 |
ENSMUST00000033923.7
|
Dlc1
|
deleted in liver cancer 1 |
| chr4_-_24851079 | 0.25 |
ENSMUST00000084781.5
ENSMUST00000108218.3 |
Klhl32
|
kelch-like 32 |
| chr5_-_24527276 | 0.25 |
ENSMUST00000088311.4
|
Gbx1
|
gastrulation brain homeobox 1 |
| chr13_-_95891905 | 0.25 |
ENSMUST00000068603.6
|
Iqgap2
|
IQ motif containing GTPase activating protein 2 |
| chr4_+_42158092 | 0.24 |
ENSMUST00000098122.2
|
Gm13306
|
predicted gene 13306 |
| chr19_+_48206025 | 0.23 |
ENSMUST00000078880.5
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr6_-_35308110 | 0.23 |
ENSMUST00000031868.4
|
Slc13a4
|
solute carrier family 13 (sodium/sulfate symporters), member 4 |
| chr5_-_125341043 | 0.23 |
ENSMUST00000111390.1
ENSMUST00000086075.6 |
Scarb1
|
scavenger receptor class B, member 1 |
| chr8_+_87472805 | 0.23 |
ENSMUST00000180700.2
ENSMUST00000182174.1 ENSMUST00000181159.1 |
Gm2694
|
predicted gene 2694 |
| chr8_+_87472838 | 0.23 |
ENSMUST00000180806.2
|
Gm2694
|
predicted gene 2694 |
| chr3_+_133310093 | 0.22 |
ENSMUST00000029644.9
ENSMUST00000122334.1 |
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr9_-_67539392 | 0.22 |
ENSMUST00000039662.8
|
Tln2
|
talin 2 |
| chr17_-_51179249 | 0.21 |
ENSMUST00000024717.8
|
Tbc1d5
|
TBC1 domain family, member 5 |
| chr19_-_60468304 | 0.21 |
ENSMUST00000051277.2
|
Prlhr
|
prolactin releasing hormone receptor |
| chr13_-_95223451 | 0.21 |
ENSMUST00000159608.1
|
Pde8b
|
phosphodiesterase 8B |
| chr7_+_45873127 | 0.21 |
ENSMUST00000107718.1
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr11_-_69920892 | 0.21 |
ENSMUST00000152589.1
ENSMUST00000108612.1 ENSMUST00000108611.1 |
Eif5a
|
eukaryotic translation initiation factor 5A |
| chr8_+_92961027 | 0.20 |
ENSMUST00000072939.6
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr1_-_166238725 | 0.20 |
ENSMUST00000038782.3
|
Mael
|
maelstrom homolog (Drosophila) |
| chr11_+_69632927 | 0.20 |
ENSMUST00000018909.3
|
Fxr2
|
fragile X mental retardation, autosomal homolog 2 |
| chr10_-_61979073 | 0.20 |
ENSMUST00000105453.1
ENSMUST00000105452.2 ENSMUST00000105454.2 |
Col13a1
|
collagen, type XIII, alpha 1 |
| chr12_-_56535047 | 0.20 |
ENSMUST00000178477.2
|
Nkx2-1
|
NK2 homeobox 1 |
| chr11_-_55608189 | 0.20 |
ENSMUST00000102716.3
|
Glra1
|
glycine receptor, alpha 1 subunit |
| chr19_+_41482632 | 0.19 |
ENSMUST00000067795.5
|
Lcor
|
ligand dependent nuclear receptor corepressor |
| chr7_-_57387172 | 0.19 |
ENSMUST00000068911.6
|
Gabrg3
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 3 |
| chr17_+_24669730 | 0.19 |
ENSMUST00000047179.5
|
Zfp598
|
zinc finger protein 598 |
| chr4_+_117096049 | 0.18 |
ENSMUST00000030443.5
|
Ptch2
|
patched homolog 2 |
| chr18_-_80713062 | 0.18 |
ENSMUST00000170905.1
ENSMUST00000078049.4 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr6_+_61180313 | 0.18 |
ENSMUST00000126214.1
|
Ccser1
|
coiled-coil serine rich 1 |
| chr6_-_148831395 | 0.18 |
ENSMUST00000145960.1
|
Ipo8
|
importin 8 |
| chr11_-_78497458 | 0.18 |
ENSMUST00000108287.3
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr6_-_148831448 | 0.17 |
ENSMUST00000048418.7
|
Ipo8
|
importin 8 |
| chr17_-_29347902 | 0.17 |
ENSMUST00000095427.4
ENSMUST00000118366.1 |
Mtch1
|
mitochondrial carrier homolog 1 (C. elegans) |
| chr2_+_164960809 | 0.17 |
ENSMUST00000124372.1
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr7_+_45872772 | 0.17 |
ENSMUST00000002855.5
ENSMUST00000107719.1 |
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr8_+_87473116 | 0.17 |
ENSMUST00000182650.1
ENSMUST00000182758.1 ENSMUST00000181898.1 |
Gm2694
|
predicted gene 2694 |
| chr9_-_63146980 | 0.16 |
ENSMUST00000055281.7
ENSMUST00000119146.1 |
Skor1
|
SKI family transcriptional corepressor 1 |
| chr12_-_75177325 | 0.15 |
ENSMUST00000042299.2
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr11_-_70687917 | 0.15 |
ENSMUST00000108545.2
ENSMUST00000120261.1 ENSMUST00000036299.7 ENSMUST00000119120.1 ENSMUST00000100933.3 |
Camta2
|
calmodulin binding transcription activator 2 |
| chr14_+_26693267 | 0.15 |
ENSMUST00000022433.4
|
Dnah12
|
dynein, axonemal, heavy chain 12 |
| chr11_+_93996082 | 0.14 |
ENSMUST00000041956.7
|
Spag9
|
sperm associated antigen 9 |
| chr3_-_95015214 | 0.14 |
ENSMUST00000128438.1
ENSMUST00000149747.1 ENSMUST00000019482.1 |
Zfp687
|
zinc finger protein 687 |
| chr7_-_47133395 | 0.14 |
ENSMUST00000107617.1
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr13_+_73260473 | 0.14 |
ENSMUST00000022095.8
|
Irx4
|
Iroquois related homeobox 4 (Drosophila) |
| chr2_-_144332146 | 0.14 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr1_-_12991109 | 0.14 |
ENSMUST00000115403.2
ENSMUST00000115402.1 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr5_-_72974702 | 0.13 |
ENSMUST00000043711.8
|
Gm10135
|
predicted gene 10135 |
| chr11_+_75733037 | 0.13 |
ENSMUST00000131398.1
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| chr11_-_89538556 | 0.13 |
ENSMUST00000169201.1
|
Ankfn1
|
ankyrin-repeat and fibronectin type III domain containing 1 |
| chr16_+_16213318 | 0.13 |
ENSMUST00000162150.1
ENSMUST00000161342.1 ENSMUST00000039408.2 |
Pkp2
|
plakophilin 2 |
| chr2_-_181288016 | 0.13 |
ENSMUST00000049032.6
|
Gmeb2
|
glucocorticoid modulatory element binding protein 2 |
| chr11_+_75732869 | 0.13 |
ENSMUST00000067664.3
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| chr4_+_130792513 | 0.12 |
ENSMUST00000070478.3
|
Sdc3
|
syndecan 3 |
| chr9_-_105495037 | 0.11 |
ENSMUST00000176190.1
ENSMUST00000163879.2 ENSMUST00000112558.2 ENSMUST00000176390.1 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr9_-_105495130 | 0.11 |
ENSMUST00000038118.7
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr17_-_80373541 | 0.11 |
ENSMUST00000086549.1
|
Gm10190
|
predicted gene 10190 |
| chr9_+_65346066 | 0.11 |
ENSMUST00000048184.2
|
Pdcd7
|
programmed cell death 7 |
| chrX_+_71364745 | 0.10 |
ENSMUST00000114601.1
ENSMUST00000146213.1 ENSMUST00000015358.1 |
Mtmr1
|
myotubularin related protein 1 |
| chr17_+_37045963 | 0.10 |
ENSMUST00000025338.9
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr3_-_89913144 | 0.10 |
ENSMUST00000029559.6
|
Il6ra
|
interleukin 6 receptor, alpha |
| chr17_-_83631892 | 0.10 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr8_+_72240315 | 0.10 |
ENSMUST00000126885.1
|
Ap1m1
|
adaptor-related protein complex AP-1, mu subunit 1 |
| chr19_-_12501996 | 0.10 |
ENSMUST00000045521.7
|
Dtx4
|
deltex 4 homolog (Drosophila) |
| chr7_-_47133684 | 0.10 |
ENSMUST00000102626.1
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr3_-_110142996 | 0.09 |
ENSMUST00000156177.2
|
Ntng1
|
netrin G1 |
| chr6_+_97807014 | 0.09 |
ENSMUST00000043637.7
|
Mitf
|
microphthalmia-associated transcription factor |
| chr5_-_142509653 | 0.09 |
ENSMUST00000110784.1
|
Radil
|
Ras association and DIL domains |
| chr11_+_93099284 | 0.09 |
ENSMUST00000092780.3
ENSMUST00000107863.2 |
Car10
|
carbonic anhydrase 10 |
| chr5_-_143138200 | 0.09 |
ENSMUST00000164536.2
|
Olfr718-ps1
|
olfactory receptor 718, pseudogene 1 |
| chr14_+_25980039 | 0.09 |
ENSMUST00000173155.1
|
Duxbl1
|
double homeobox B-like 1 |
| chrX_-_111697069 | 0.09 |
ENSMUST00000113422.2
ENSMUST00000038472.5 |
Hdx
|
highly divergent homeobox |
| chr17_+_37045980 | 0.08 |
ENSMUST00000174456.1
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr1_+_10039762 | 0.07 |
ENSMUST00000122156.1
ENSMUST00000118263.1 ENSMUST00000119714.1 |
Cspp1
|
centrosome and spindle pole associated protein 1 |
| chr4_-_130175113 | 0.07 |
ENSMUST00000105998.1
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr1_+_153665587 | 0.07 |
ENSMUST00000147700.1
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_62360524 | 0.07 |
ENSMUST00000107461.1
ENSMUST00000084528.3 |
Fkbp15
|
FK506 binding protein 15 |
| chr2_+_180499893 | 0.07 |
ENSMUST00000029084.2
|
Ntsr1
|
neurotensin receptor 1 |
| chr16_+_57353271 | 0.06 |
ENSMUST00000099667.2
|
Filip1l
|
filamin A interacting protein 1-like |
| chr3_+_107595031 | 0.06 |
ENSMUST00000014747.1
|
Alx3
|
aristaless-like homeobox 3 |
| chr2_-_167661496 | 0.06 |
ENSMUST00000125544.2
ENSMUST00000006587.6 |
Gm20431
Tmem189
|
predicted gene 20431 transmembrane protein 189 |
| chr5_+_100845709 | 0.06 |
ENSMUST00000144623.1
|
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9 |
| chr1_+_153665627 | 0.06 |
ENSMUST00000147482.1
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr3_-_83129246 | 0.06 |
ENSMUST00000047876.6
|
Gm10710
|
predicted gene 10710 |
| chr2_-_161109017 | 0.05 |
ENSMUST00000039782.7
ENSMUST00000134178.1 |
Chd6
|
chromodomain helicase DNA binding protein 6 |
| chr1_+_192190771 | 0.05 |
ENSMUST00000078470.5
ENSMUST00000110844.1 |
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1 |
| chr11_-_70669283 | 0.05 |
ENSMUST00000129434.1
ENSMUST00000018431.6 |
Spag7
|
sperm associated antigen 7 |
| chr4_-_108833608 | 0.04 |
ENSMUST00000102742.4
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr19_-_57008187 | 0.04 |
ENSMUST00000118800.1
ENSMUST00000111584.2 ENSMUST00000122359.1 ENSMUST00000148049.1 |
Afap1l2
|
actin filament associated protein 1-like 2 |
| chr7_-_127026479 | 0.04 |
ENSMUST00000032916.4
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_116335384 | 0.04 |
ENSMUST00000036215.7
|
Foxj1
|
forkhead box J1 |
| chr14_+_26119811 | 0.04 |
ENSMUST00000173617.1
|
Duxbl2
|
doubl homeobox B-like 2 |
| chrX_-_74085586 | 0.04 |
ENSMUST00000123362.1
ENSMUST00000140399.1 ENSMUST00000100750.3 |
Mecp2
|
methyl CpG binding protein 2 |
| chr7_+_27653906 | 0.04 |
ENSMUST00000008088.7
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr4_+_128883549 | 0.04 |
ENSMUST00000035667.8
|
Trim62
|
tripartite motif-containing 62 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.7 | GO:0051410 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.4 | 1.1 | GO:0097274 | urea homeostasis(GO:0097274) |
| 0.3 | 0.9 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.2 | 0.9 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 0.7 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.2 | 0.6 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.2 | 1.1 | GO:0097473 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.2 | 0.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 1.3 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.1 | 0.7 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.6 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.4 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.6 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.5 | GO:0061428 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.4 | GO:0097490 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) sympathetic neuron projection extension(GO:0097490) sympathetic neuron projection guidance(GO:0097491) |
| 0.1 | 0.7 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.1 | 0.4 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) mesenchymal stem cell proliferation(GO:0097168) |
| 0.1 | 0.5 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.1 | 0.3 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.3 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.3 | GO:1903588 | blood vessel lumenization(GO:0072554) negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.1 | 1.4 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.9 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.2 | GO:0015920 | regulation of phosphatidylcholine catabolic process(GO:0010899) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 1.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.6 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.1 | 1.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.1 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.3 | GO:0072719 | cellular response to cisplatin(GO:0072719) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.2 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.5 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 0.2 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.2 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.3 | GO:0070317 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) negative regulation of G0 to G1 transition(GO:0070317) |
| 0.0 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.3 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 1.5 | GO:0036075 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 1.6 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.0 | 0.3 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.7 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.0 | 0.2 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.4 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.5 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.2 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.4 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.0 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.0 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.2 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.3 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.3 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.4 | GO:2000279 | negative regulation of DNA biosynthetic process(GO:2000279) |
| 0.0 | 0.3 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.8 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 0.6 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.5 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.1 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.4 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 1.0 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.2 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.5 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 1.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.3 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.2 | GO:0031528 | microvillus membrane(GO:0031528) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.4 | 1.2 | GO:0051765 | inositol tetrakisphosphate kinase activity(GO:0051765) |
| 0.2 | 0.7 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 0.6 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.2 | 0.5 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 1.4 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 3.7 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.1 | 0.5 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.1 | GO:0004915 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.1 | 0.7 | GO:0002054 | nucleobase binding(GO:0002054) hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.3 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.1 | 1.1 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.7 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.9 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) hedgehog receptor activity(GO:0008158) |
| 0.1 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.1 | 0.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.2 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.1 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.1 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.7 | GO:0010314 | DNA binding, bending(GO:0008301) phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.8 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 1.9 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.3 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.3 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.3 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.4 | GO:0045499 | neuropilin binding(GO:0038191) chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.7 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.8 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.2 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.2 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.9 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.4 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 2.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.7 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.7 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.5 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |