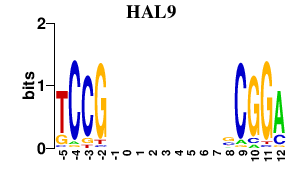
Results for HAL9
Z-value: 0.73
Transcription factors associated with HAL9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HAL9
|
S000005449 | Putative transcription factor containing a zinc finger |
Activity-expression correlation:
Activity profile of HAL9 motif
Sorted Z-values of HAL9 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YJR094W-A | 5.82 |
RPL43B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl43Ap and has similarity to rat L37a ribosomal protein |
|
| YHR216W | 4.74 |
IMD2
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, expression is induced by mycophenolic acid resulting in resistance to the drug, expression is repressed by nutrient limitation |
|
| YLR154W-A | 4.58 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154W-B | 4.46 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YLR154C | 4.39 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YIR021W | 4.11 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YPL250W-A | 3.98 |
Identified by fungal homology and RT-PCR |
||
| YKR092C | 3.92 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YLL045C | 3.61 |
RPL8B
|
Ribosomal protein L4 of the large (60S) ribosomal subunit, nearly identical to Rpl8Ap and has similarity to rat L7a ribosomal protein; mutation results in decreased amounts of free 60S subunits |
|
| YGL209W | 3.61 |
MIG2
|
Protein containing zinc fingers, involved in repression, along with Mig1p, of SUC2 (invertase) expression by high levels of glucose; binds to Mig1p-binding sites in SUC2 promoter |
|
| YLR154W-C | 3.29 |
TAR1
|
Mitochondrial protein of unknown function, overexpression suppresses an rpo41 mutation affecting mitochondrial RNA polymerase; encoded within the 25S rRNA gene on the opposite strand |
|
| YDL075W | 3.16 |
RPL31A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl31Bp and has similarity to rat L31 ribosomal protein; associates with the karyopherin Sxm1p |
|
| YLR349W | 3.02 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified ORF DIC1/YLR348C |
||
| YPR148C | 2.90 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YLR348C | 2.86 |
DIC1
|
Mitochondrial dicarboxylate carrier, integral membrane protein, catalyzes a dicarboxylate-phosphate exchange across the inner mitochondrial membrane, transports cytoplasmic dicarboxylates into the mitochondrial matrix |
|
| YGR280C | 2.79 |
PXR1
|
Essential protein involved in rRNA and snoRNA maturation; competes with TLC1 RNA for binding to Est2p, suggesting a role in negative regulation of telomerase; human homolog inhibits telomerase; contains a G-patch RNA interacting domain |
|
| YNR013C | 2.78 |
PHO91
|
Low-affinity phosphate transporter of the vacuolar membrane; deletion of pho84, pho87, pho89, pho90, and pho91 causes synthetic lethality; transcription independent of Pi and Pho4p activity; overexpression results in vigorous growth |
|
| YLR301W | 2.77 |
Protein of unknown function that interacts with Sec72p |
||
| YOR101W | 2.77 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YNL178W | 2.74 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YJL136C | 2.65 |
RPS21B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps21Ap and has similarity to rat S21 ribosomal protein |
|
| YFL022C | 2.63 |
FRS2
|
Alpha subunit of cytoplasmic phenylalanyl-tRNA synthetase, forms a tetramer with Frs1p to form active enzyme; evolutionarily distant from mitochondrial phenylalanyl-tRNA synthetase based on protein sequence, but substrate binding is similar |
|
| YJL153C | 2.51 |
INO1
|
Inositol 1-phosphate synthase, involved in synthesis of inositol phosphates and inositol-containing phospholipids; transcription is coregulated with other phospholipid biosynthetic genes by Ino2p and Ino4p, which bind the UASINO DNA element |
|
| YLR413W | 2.33 |
Putative protein of unknown function; YLR413W is not an essential gene |
||
| YIL056W | 2.28 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YHR141C | 2.26 |
RPL42B
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl42Ap and has similarity to rat L44; required for propagation of the killer toxin-encoding M1 double-stranded RNA satellite of the L-A double-stranded RNA virus |
|
| YCL024W | 2.23 |
KCC4
|
Protein kinase of the bud neck involved in the septin checkpoint, associates with septin proteins, negatively regulates Swe1p by phosphorylation, shows structural homology to bud neck kinases Gin4p and Hsl1p |
|
| YDR037W | 2.23 |
KRS1
|
Lysyl-tRNA synthetase; also identified as a negative regulator of general control of amino acid biosynthesis |
|
| YKL096W-A | 2.20 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YPL249C-A | 2.13 |
RPL36B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl36Ap and has similarity to rat L36 ribosomal protein; binds to 5.8 S rRNA |
|
| YDR497C | 2.07 |
ITR1
|
Myo-inositol transporter with strong similarity to the minor myo-inositol transporter Itr2p, member of the sugar transporter superfamily; expression is repressed by inositol and choline via Opi1p and derepressed via Ino2p and Ino4p |
|
| YDR418W | 1.99 |
RPL12B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl12Ap; rpl12a rpl12b double mutant exhibits slow growth and slow translation; has similarity to E. coli L11 and rat L12 ribosomal proteins |
|
| YMR108W | 1.99 |
ILV2
|
Acetolactate synthase, catalyses the first common step in isoleucine and valine biosynthesis and is the target of several classes of inhibitors, localizes to the mitochondria; expression of the gene is under general amino acid control |
|
| YMR102C | 1.96 |
Protein of unknown function; transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in multidrug resistance; mutant shows increased resistance to azoles; YMR102C is not an essential gene |
||
| YLR185W | 1.92 |
RPL37A
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl37Bp and to rat L37 ribosomal protein |
|
| YCL023C | 1.91 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF KCC4 |
||
| YIL133C | 1.89 |
RPL16A
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Bp, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YLR183C | 1.74 |
TOS4
|
Transcription factor that binds to a number of promoter regions, particularly promoters of some genes involved in pheromone response and cell cycle; potential Cdc28p substrate; expression is induced in G1 by bound SBF |
|
| YNL153C | 1.73 |
GIM3
|
Subunit of the heterohexameric cochaperone prefoldin complex which binds specifically to cytosolic chaperonin and transfers target proteins to it |
|
| YMR009W | 1.71 |
ADI1
|
Acireductone dioxygenease involved in the methionine salvage pathway; ortholog of human MTCBP-1; transcribed with YMR010W and regulated post-transcriptionally by RNase III (Rnt1p) cleavage; ADI1 mRNA is induced in heat shock conditions |
|
| YPR074C | 1.69 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YDL082W | 1.69 |
RPL13A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl13Bp; not essential for viability; has similarity to rat L13 ribosomal protein |
|
| YOL127W | 1.63 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YIL118W | 1.61 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YDL083C | 1.60 |
RPS16B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps16Ap and has similarity to E. coli S9 and rat S16 ribosomal proteins |
|
| YML024W | 1.60 |
RPS17A
|
Ribosomal protein 51 (rp51) of the small (40s) subunit; nearly identical to Rps17Bp and has similarity to rat S17 ribosomal protein |
|
| YGL201C | 1.58 |
MCM6
|
Protein involved in DNA replication; component of the Mcm2-7 hexameric complex that binds chromatin as a part of the pre-replicative complex |
|
| YFL015W-A | 1.57 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YPL062W | 1.54 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YPL062W is not an essential gene; homozygous diploid mutant shows a decrease in glycogen accumulation |
||
| YGR138C | 1.54 |
TPO2
|
Polyamine transport protein specific for spermine; localizes to the plasma membrane; transcription of TPO2 is regulated by Haa1p; member of the major facilitator superfamily |
|
| YKL218C | 1.54 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YPL137C | 1.53 |
GIP3
|
Glc7-interacting protein whose overexpression relocalizes Glc7p from the nucleus and prevents chromosome segregation; may interact with ribosomes, based on co-purification experiments |
|
| YGR148C | 1.50 |
RPL24B
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Ap and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YIL121W | 1.48 |
QDR2
|
Multidrug transporter of the major facilitator superfamily, required for resistance to quinidine, barban, cisplatin, and bleomycin; may have a role in potassium uptake |
|
| YNL090W | 1.48 |
RHO2
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, involved in the establishment of cell polarity and in microtubule assembly |
|
| YFL015C | 1.46 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YMR099C | 1.46 |
Glucose-6-phosphate 1-epimerase (hexose-6-phosphate mutarotase), likely involved in carbohydrate metabolism; GFP-fusion protein localizes to both the nucleus and cytoplasm and is induced in response to the DNA-damaging agent MMS |
||
| YER146W | 1.46 |
LSM5
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YNL069C | 1.42 |
RPL16B
|
N-terminally acetylated protein component of the large (60S) ribosomal subunit, binds to 5.8 S rRNA; has similarity to Rpl16Ap, E. coli L13 and rat L13a ribosomal proteins; transcriptionally regulated by Rap1p |
|
| YHR136C | 1.41 |
SPL2
|
Protein with similarity to cyclin-dependent kinase inhibitors, overproduction suppresses a plc1 null mutation; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
|
| YJL190C | 1.40 |
RPS22A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps22Bp and has similarity to E. coli S8 and rat S15a ribosomal proteins |
|
| YKL120W | 1.38 |
OAC1
|
Mitochondrial inner membrane transporter, transports oxaloacetate, sulfate, and thiosulfate; member of the mitochondrial carrier family |
|
| YKR093W | 1.36 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YLR048W | 1.32 |
RPS0B
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps0Ap; required for maturation of 18S rRNA along with Rps0Ap; deletion of either RPS0 gene reduces growth rate, deletion of both genes is lethal |
|
| YOL101C | 1.32 |
IZH4
|
Membrane protein involved in zinc metabolism, member of the four-protein IZH family, expression induced by fatty acids and altered zinc levels; deletion reduces sensitivity to excess zinc; possible role in sterol metabolism |
|
| YGR097W | 1.32 |
ASK10
|
Component of the RNA polymerase II holoenzyme, phosphorylated in response to oxidative stress; has a role in destruction of Ssn8p, which relieves repression of stress-response genes |
|
| YMR121C | 1.31 |
RPL15B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl15Ap and has similarity to rat L15 ribosomal protein; binds to 5.8 S rRNA |
|
| YNR016C | 1.31 |
ACC1
|
Acetyl-CoA carboxylase, biotin containing enzyme that catalyzes the carboxylation of acetyl-CoA to form malonyl-CoA; required for de novo biosynthesis of long-chain fatty acids |
|
| YGR242W | 1.30 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF YAP1802/YGR241C |
||
| YKR074W | 1.30 |
AIM29
|
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm; null mutant displays increased frequency of mitochondrial genome loss (petite formation) |
|
| YOR340C | 1.25 |
RPA43
|
RNA polymerase I subunit A43 |
|
| YFR055W | 1.25 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YOR163W | 1.23 |
DDP1
|
Polyphosphate phosphatase; hydrolyzes diphosphorylated inositol polyphosphates and diadenosine polyphosphates; has high specificity for diadenosine hexa- and pentaphosphates; member of the MutT family of nucleotide hydrolases |
|
| YMR208W | 1.22 |
ERG12
|
Mevalonate kinase, acts in the biosynthesis of isoprenoids and sterols, including ergosterol, from mevalonate |
|
| YGR140W | 1.20 |
CBF2
|
Essential kinetochore protein, component of the CBF3 multisubunit complex that binds to the CDEIII region of the centromere; Cbf2p also binds to the CDEII region possibly forming a different multimeric complex, ubiquitinated in vivo |
|
| YDR144C | 1.19 |
MKC7
|
GPI-anchored aspartyl protease (yapsin) involved in protein processing; shares functions with Yap3p and Kex2p |
|
| YJL212C | 1.17 |
OPT1
|
Proton-coupled oligopeptide transporter of the plasma membrane; also transports glutathione and phytochelatin; member of the OPT family |
|
| YHR137W | 1.17 |
ARO9
|
Aromatic aminotransferase II, catalyzes the first step of tryptophan, phenylalanine, and tyrosine catabolism |
|
| YGR241C | 1.17 |
YAP1802
|
Protein involved in clathrin cage assembly; binds Pan1p and clathrin; homologous to Yap1801p, member of the AP180 protein family |
|
| YPR063C | 1.16 |
ER-localized protein of unknown function |
||
| YGR118W | 1.15 |
RPS23A
|
Ribosomal protein 28 (rp28) of the small (40S) ribosomal subunit, required for translational accuracy; nearly identical to Rps23Bp and similar to E. coli S12 and rat S23 ribosomal proteins; deletion of both RPS23A and RPS23B is lethal |
|
| YOR234C | 1.13 |
RPL33B
|
Ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Ap and has similarity to rat L35a; rpl33b null mutant exhibits normal growth while rpl33a rpl33b double null mutant is inviable |
|
| YHR020W | 1.12 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; has similarity to proline-tRNA ligase; YHR020W is an essential gene |
||
| YGR034W | 1.11 |
RPL26B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Ap and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YDR344C | 1.11 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGR128C | 1.08 |
UTP8
|
Nucleolar protein required for export of tRNAs from the nucleus; also copurifies with the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YHR094C | 1.07 |
HXT1
|
Low-affinity glucose transporter of the major facilitator superfamily, expression is induced by Hxk2p in the presence of glucose and repressed by Rgt1p when glucose is limiting |
|
| YFR056C | 1.07 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YMR100W | 1.06 |
MUB1
|
Protein of unknown function, deletion causes multi-budding phenotype; has similarity to Aspergillus nidulans samB gene |
|
| YLR333C | 1.05 |
RPS25B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps25Ap and has similarity to rat S25 ribosomal protein |
|
| YGR139W | 0.99 |
Dubious ORF unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL035C | 0.99 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YDR215C | 0.99 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; null mutant displays elevated sensitivity to expression of a mutant huntingtin fragment or of alpha-synuclein |
||
| YDL241W | 0.97 |
Putative protein of unknown function; YDL241W is not an essential gene |
||
| YNL089C | 0.97 |
Dubious open reading frame unlikely to encode a functional protein; almost completely overlaps YNL090W/RHO2 which encodes a small GTPase of the Rho/Rac subfamily of Ras-like proteins |
||
| YMR267W | 0.96 |
PPA2
|
Mitochondrial inorganic pyrophosphatase, required for mitochondrial function and possibly involved in energy generation from inorganic pyrophosphate |
|
| YGR017W | 0.96 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the cytoplasm |
||
| YCR099C | 0.96 |
Putative protein of unknown function |
||
| YKR094C | 0.96 |
RPL40B
|
Fusion protein, identical to Rpl40Ap, that is cleaved to yield ubiquitin and a ribosomal protein of the large (60S) ribosomal subunit with similarity to rat L40; ubiquitin may facilitate assembly of the ribosomal protein into ribosomes |
|
| YGR052W | 0.94 |
FMP48
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; induced by treatment with 8-methoxypsoralen and UVA irradiation |
|
| YPL170W | 0.94 |
DAP1
|
Heme-binding protein involved in regulation of cytochrome P450 protein Erg11p; damage response protein, related to mammalian membrane progesterone receptors; mutations lead to defects in telomeres, mitochondria, and sterol synthesis |
|
| YPL136W | 0.94 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified gene GIP3/YPL137C |
||
| YIL039W | 0.93 |
TED1
|
Conserved phosphoesterase domain-containing protein that acts together with Emp24p/Erv25p in cargo exit from the ER; deletion confers sensitivity to 4-(N-(S-glutathionylacetyl)amino) phenylarsenoxide (GSAO) |
|
| YLL028W | 0.92 |
TPO1
|
Polyamine transporter that recognizes spermine, putrescine, and spermidine; catalyzes uptake of polyamines at alkaline pH and excretion at acidic pH; phosphorylation enhances activity and sorting to the plasma membrane |
|
| YLR025W | 0.91 |
SNF7
|
One of four subunits of the endosomal sorting complex required for transport III (ESCRT-III); involved in the sorting of transmembrane proteins into the multivesicular body (MVB) pathway; recruited from the cytoplasm to endosomal membranes |
|
| YLR448W | 0.89 |
RPL6B
|
Protein component of the large (60S) ribosomal subunit, has similarity to Rpl6Ap and to rat L6 ribosomal protein; binds to 5.8S rRNA |
|
| YJR094C | 0.89 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YLR262C-A | 0.88 |
TMA7
|
Protein of unknown that associates with ribosomes; null mutant exhibits translation defects, altered polyribosome profiles, and resistance to the translation inhibitor anisomcyin |
|
| YJL222W | 0.87 |
VTH2
|
Putative membrane glycoprotein with strong similarity to Vth1p and Pep1p/Vps10p, may be involved in vacuolar protein sorting |
|
| YML063W | 0.87 |
RPS1B
|
Ribosomal protein 10 (rp10) of the small (40S) subunit; nearly identical to Rps1Ap and has similarity to rat S3a ribosomal protein |
|
| YBR106W | 0.86 |
PHO88
|
Probable membrane protein, involved in phosphate transport; pho88 pho86 double null mutant exhibits enhanced synthesis of repressible acid phosphatase at high inorganic phosphate concentrations |
|
| YGR055W | 0.85 |
MUP1
|
High affinity methionine permease, integral membrane protein with 13 putative membrane-spanning regions; also involved in cysteine uptake |
|
| YBR118W | 0.85 |
TEF2
|
Translational elongation factor EF-1 alpha; also encoded by TEF1; functions in the binding reaction of aminoacyl-tRNA (AA-tRNA) to ribosomes |
|
| YDL023C | 0.85 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YKL106W | 0.84 |
AAT1
|
Mitochondrial aspartate aminotransferase, catalyzes the conversion of oxaloacetate to aspartate in aspartate and asparagine biosynthesis |
|
| YER036C | 0.83 |
ARB1
|
ATPase of the ATP-binding cassette (ABC) family involved in 40S and 60S ribosome biogenesis, has similarity to Gcn20p; shuttles from nucleus to cytoplasm, physically interacts with Tif6p, Lsg1p |
|
| YKL065C | 0.81 |
YET1
|
Endoplasmic reticulum transmembrane protein; may interact with ribosomes, based on co-purification experiments; homolog of human BAP31 protein |
|
| YKL068W-A | 0.81 |
Putative protein of unknown function; identified by homology to Ashbya gossypii |
||
| YGR050C | 0.80 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR158W | 0.80 |
AMN1
|
Protein required for daughter cell separation, multiple mitotic checkpoints, and chromosome stability; contains 12 degenerate leucine-rich repeat motifs; expression is induced by the Mitotic Exit Network (MEN) |
|
| YHR019C | 0.79 |
DED81
|
Cytosolic asparaginyl-tRNA synthetase, required for protein synthesis, catalyzes the specific attachment of asparagine to its cognate tRNA |
|
| YML088W | 0.79 |
UFO1
|
F-box receptor protein, subunit of the Skp1-Cdc53-F-box receptor (SCF) E3 ubiquitin ligase complex; binds to phosphorylated Ho endonuclease, allowing its ubiquitylation by SCF and subsequent degradation |
|
| YJR123W | 0.77 |
RPS5
|
Protein component of the small (40S) ribosomal subunit, the least basic of the non-acidic ribosomal proteins; phosphorylated in vivo; essential for viability; has similarity to E. coli S7 and rat S5 ribosomal proteins |
|
| YLR344W | 0.76 |
RPL26A
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl26Bp and has similarity to E. coli L24 and rat L26 ribosomal proteins; binds to 5.8S rRNA |
|
| YER001W | 0.76 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YFR036W-A | 0.76 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene RSC8 |
||
| YLL043W | 0.75 |
FPS1
|
Plasma membrane channel, member of major intrinsic protein (MIP) family; involved in efflux of glycerol and in uptake of acetic acid and the trivalent metalloids arsenite and antimonite; phosphorylated by Hog1p MAPK under acetate stress |
|
| YFL016C | 0.75 |
MDJ1
|
Co-chaperone that stimulates the ATPase activity of the HSP70 protein Ssc1p; involved in protein folding/refolding in the mitochodrial matrix; required for proteolysis of misfolded proteins; member of the HSP40 (DnaJ) family of chaperones |
|
| YDR072C | 0.74 |
IPT1
|
Inositolphosphotransferase 1, involved in synthesis of mannose-(inositol-P)2-ceramide (M(IP)2C), which is the most abundant sphingolipid in cells, mutation confers resistance to the antifungals syringomycin E and DmAMP1 in some growth media |
|
| YDL022W | 0.73 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YHR007C | 0.72 |
ERG11
|
Lanosterol 14-alpha-demethylase, catalyzes the C-14 demethylation of lanosterol to form 4,4''-dimethyl cholesta-8,14,24-triene-3-beta-ol in the ergosterol biosynthesis pathway; member of the cytochrome P450 family |
|
| YKL110C | 0.72 |
KTI12
|
Protein that plays a role, with Elongator complex, in modification of wobble nucleosides in tRNA; involved in sensitivity to G1 arrest induced by zymocin; interacts with chromatin throughout the genome; also interacts with Cdc19p |
|
| YIR013C | 0.72 |
GAT4
|
Protein containing GATA family zinc finger motifs |
|
| YOR119C | 0.71 |
RIO1
|
Essential serine kinase involved in cell cycle progression and processing of the 20S pre-rRNA into mature 18S rRNA |
|
| YPL004C | 0.69 |
LSP1
|
Primary component of eisosomes, which are large immobile patch structures at the cell cortex associated with endocytosis, along with Pil1p and Sur7p; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways |
|
| YGL034C | 0.67 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YCR018C | 0.66 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YDL137W | 0.66 |
ARF2
|
ADP-ribosylation factor, GTPase of the Ras superfamily involved in regulation of coated formation vesicles in intracellular trafficking within the Golgi; functionally interchangeable with Arf1p |
|
| YMR122W-A | 0.66 |
Protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and endoplasmic reticulum |
||
| YML126C | 0.65 |
ERG13
|
3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase, catalyzes the formation of HMG-CoA from acetyl-CoA and acetoacetyl-CoA; involved in the second step in mevalonate biosynthesis |
|
| YJR047C | 0.65 |
ANB1
|
Translation initiation factor eIF-5A, promotes formation of the first peptide bond; similar to and functionally redundant with Hyp2p; undergoes an essential hypusination modification; expressed under anaerobic conditions |
|
| YPL131W | 0.64 |
RPL5
|
Protein component of the large (60S) ribosomal subunit with similarity to E. coli L18 and rat L5 ribosomal proteins; binds 5S rRNA and is required for 60S subunit assembly |
|
| YKR038C | 0.64 |
KAE1
|
Putative glycoprotease proposed to be in transcription as a component of the EKC protein complex with Bud32p, Cgi121p, Pcc1p, and Gon7p; also identified as a component of the KEOPS protein complex |
|
| YKL063C | 0.64 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the Golgi |
||
| YNL189W | 0.63 |
SRP1
|
Karyopherin alpha homolog, forms a dimer with karyopherin beta Kap95p to mediate import of nuclear proteins, binds the nuclear localization signal of the substrate during import; may also play a role in regulation of protein degradation |
|
| YNL304W | 0.63 |
YPT11
|
Rab-type small GTPase that interacts with the C-terminal tail domain of Myo2p to mediate distribution of mitochondria to daughter cells |
|
| YLL027W | 0.63 |
ISA1
|
Mitochondrial matrix protein involved in biogenesis of the iron-sulfur (Fe/S) cluster of Fe/S proteins, isa1 deletion causes loss of mitochondrial DNA and respiratory deficiency; depletion reduces growth on nonfermentable carbon sources |
|
| YOR276W | 0.63 |
CAF20
|
Phosphoprotein of the mRNA cap-binding complex involved in translational control, repressor of cap-dependent translation initiation, competes with eIF4G for binding to eIF4E |
|
| YJL223C | 0.62 |
PAU1
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YPL003W | 0.62 |
ULA1
|
Protein that acts together with Uba3p to activate Rub1p before its conjugation to proteins (neddylation), which may play a role in protein degradation |
|
| YJL173C | 0.62 |
RFA3
|
Subunit of heterotrimeric Replication Protein A (RPA), which is a highly conserved single-stranded DNA binding protein involved in DNA replication, repair, and recombination |
|
| YJL115W | 0.61 |
ASF1
|
Nucleosome assembly factor, involved in chromatin assembly and disassembly, anti-silencing protein that causes derepression of silent loci when overexpressed; plays a role in regulating Ty1 transposition |
|
| YDR451C | 0.61 |
YHP1
|
One of two homeobox transcriptional repressors (see also Yox1p), that bind to Mcm1p and to early cell cycle box (ECB) elements of cell cycle regulated genes, thereby restricting ECB-mediated transcription to the M/G1 interval |
|
| YBL032W | 0.61 |
HEK2
|
RNA binding protein with similarity to hnRNP-K that localizes to the cytoplasm and to subtelomeric DNA; required for the proper localization of ASH1 mRNA; involved in the regulation of telomere position effect and telomere length |
|
| YEL020C-B | 0.61 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene YEL020W-A; identified by fungal homology and RT-PCR |
||
| YPR074W-A | 0.60 |
Hypothetical protein identified by homology |
||
| YIR019C | 0.60 |
MUC1
|
GPI-anchored cell surface glycoprotein (flocculin) required for pseudohyphal formation, invasive growth, flocculation, and biofilms; transcriptionally regulated by the MAPK pathway (via Ste12p and Tec1p) and the cAMP pathway (via Flo8p) |
|
| YKL209C | 0.60 |
STE6
|
Plasma membrane ATP-binding cassette (ABC) transporter required for the export of a-factor, catalyzes ATP hydrolysis coupled to a-factor transport; contains 12 transmembrane domains and two ATP binding domains; expressed only in MATa cells |
|
| YNL302C | 0.59 |
RPS19B
|
Protein component of the small (40S) ribosomal subunit, required for assembly and maturation of pre-40 S particles; mutations in human RPS19 are associated with Diamond Blackfan anemia; nearly identical to Rps19Ap |
|
| YJL222W-A | 0.59 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL081C | 0.59 |
RPP1A
|
Ribosomal stalk protein P1 alpha, involved in the interaction between translational elongation factors and the ribosome; accumulation of P1 in the cytoplasm is regulated by phosphorylation and interaction with the P2 stalk component |
|
| YLR110C | 0.58 |
CCW12
|
Cell wall mannoprotein, mutants are defective in mating and agglutination, expression is downregulated by alpha-factor |
|
| YEL001C | 0.57 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YGR051C | 0.57 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YEL056W | 0.55 |
HAT2
|
Subunit of the Hat1p-Hat2p histone acetyltransferase complex; required for high affinity binding of the complex to free histone H4, thereby enhancing Hat1p activity; similar to human RbAp46 and 48; has a role in telomeric silencing |
|
| YNR028W | 0.55 |
CPR8
|
Peptidyl-prolyl cis-trans isomerase (cyclophilin), catalyzes the cis-trans isomerization of peptide bonds N-terminal to proline residues; similarity to Cpr4p suggests a potential role in the secretory pathway |
|
| YKL001C | 0.55 |
MET14
|
Adenylylsulfate kinase, required for sulfate assimilation and involved in methionine metabolism |
|
| YIL069C | 0.55 |
RPS24B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Ap and has similarity to rat S24 ribosomal protein |
|
| YHR169W | 0.54 |
DBP8
|
Putative ATP-dependent RNA helicase of the DEAD-box family involved in biogenesis of the 40S ribosomal subunit |
|
| YHR142W | 0.54 |
CHS7
|
Protein of unknown function, involved in chitin biosynthesis by regulating Chs3p export from the ER |
|
| YNL065W | 0.54 |
AQR1
|
Plasma membrane multidrug transporter of the major facilitator superfamily, confers resistance to short-chain monocarboxylic acids and quinidine; involved in the excretion of excess amino acids |
|
| YIL051C | 0.54 |
MMF1
|
Mitochondrial protein involved in maintenance of the mitochondrial genome |
|
| YDR073W | 0.54 |
SNF11
|
Subunit of the SWI/SNF chromatin remodeling complex involved in transcriptional regulation; interacts with a highly conserved 40-residue sequence of Snf2p |
|
| YDR156W | 0.54 |
RPA14
|
RNA polymerase I subunit A14 |
|
| YBR048W | 0.54 |
RPS11B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps11Ap and has similarity to E. coli S17 and rat S11 ribosomal proteins |
|
| YDL076C | 0.52 |
RXT3
|
Subunit of the RPD3L complex; involved in histone deacetylation |
|
| YIR020C | 0.51 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YOL125W | 0.51 |
TRM13
|
2'-O-methyltransferase responsible for modification of tRNA at position 4; exhibits no obvious similarity to other known methyltransferases |
|
| YGR004W | 0.50 |
PEX31
|
Peroxisomal integral membrane protein, involved in negative regulation of peroxisome size; partially functionally redundant with Pex30p and Pex32p; probably acts at a step downstream of steps mediated by Pex28p and Pex29p |
|
| YPL178W | 0.49 |
CBC2
|
Small subunit of the heterodimeric cap binding complex that also contains Sto1p, component of the spliceosomal commitment complex; interacts with Npl3p, possibly to package mRNA for export from the nucleus; contains an RNA-binding motif |
|
| YNL251C | 0.49 |
NRD1
|
RNA-binding protein that interacts with the C-terminal domain of the RNA polymerase II large subunit (Rpo21p), required for transcription termination and 3' end maturation of nonpolyadenylated RNAs |
|
| YAL038W | 0.49 |
CDC19
|
Pyruvate kinase, functions as a homotetramer in glycolysis to convert phosphoenolpyruvate to pyruvate, the input for aerobic (TCA cycle) or anaerobic (glucose fermentation) respiration |
|
| YBL063W | 0.48 |
KIP1
|
Kinesin-related motor protein required for mitotic spindle assembly and chromosome segregation; functionally redundant with Cin8p |
|
| YPL112C | 0.48 |
PEX25
|
Peripheral peroxisomal membrane peroxin required for the regulation of peroxisome size and maintenance, recruits GTPase Rho1p to peroxisomes, induced by oleate, interacts with homologous protein Pex27p |
|
| YPL263C | 0.48 |
KEL3
|
Cytoplasmic protein of unknown function |
|
| YGL031C | 0.47 |
RPL24A
|
Ribosomal protein L30 of the large (60S) ribosomal subunit, nearly identical to Rpl24Bp and has similarity to rat L24 ribosomal protein; not essential for translation but may be required for normal translation rate |
|
| YDR074W | 0.47 |
TPS2
|
Phosphatase subunit of the trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; expression is induced by stress conditions and repressed by the Ras-cAMP pathway |
|
| YML026C | 0.46 |
RPS18B
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Ap and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YEL062W | 0.46 |
NPR2
|
Protein with a possible role in regulating expression of nitrogen permeases; transcription is induced in response to proline and urea; contains two PEST sequences; null mutant is resistant to cisplatin and doxorubicin |
|
| YJL216C | 0.45 |
Protein of unknown function, similar to alpha-D-glucosidases; transcriptionally activated by both Pdr8p and Yrm1p, along with transporters and other genes involved in the pleiotropic drug resistance (PDR) phenomenon |
||
| YER045C | 0.45 |
ACA1
|
Basic leucine zipper (bZIP) transcription factor of the ATF/CREB family, may regulate transcription of genes involved in utilization of non-optimal carbon sources |
|
| YER046W | 0.45 |
SPO73
|
Meiosis-specific protein of unknown function, required for spore wall formation during sporulation; dispensible for both nuclear divisions during meiosis |
|
| YIR020W-A | 0.45 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data |
||
| YJL011C | 0.44 |
RPC17
|
RNA polymerase III subunit C17; physically interacts with C31, C11, and TFIIIB70; may be involved in the recruitment of pol III by the preinitiation complex |
|
| YOL014W | 0.44 |
Putative protein of unknown function |
||
| YFR031C-A | 0.44 |
RPL2A
|
Protein component of the large (60S) ribosomal subunit, identical to Rpl2Bp and has similarity to E. coli L2 and rat L8 ribosomal proteins |
|
| YNL118C | 0.44 |
DCP2
|
Catalytic subunit of the Dcp1p-Dcp2p decapping enzyme complex, which removes the 5' cap structure from mRNAs prior to their degradation; member of the Nudix hydrolase family |
|
| YBL006C | 0.43 |
LDB7
|
Component of the RSC chromatin remodeling complex; interacts with Rsc3p, Rsc30p, Npl6p, and Htl1p to form a module important for a broad range of RSC functions |
|
| YPR025C | 0.43 |
CCL1
|
Cyclin associated with protein kinase Kin28p, which is the TFIIH-associated carboxy-terminal domain (CTD) kinase involved in transcription initiation at RNA polymerase II promoters |
|
| YJL029C | 0.42 |
VPS53
|
Component of the GARP (Golgi-associated retrograde protein) complex, Vps51p-Vps52p-Vps53p-Vps54p, which is required for the recycling of proteins from endosomes to the late Golgi; required for vacuolar protein sorting |
Network of associatons between targets according to the STRING database.
First level regulatory network of HAL9
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0006183 | GTP biosynthetic process(GO:0006183) GTP metabolic process(GO:0046039) |
| 1.2 | 1.2 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 1.2 | 4.8 | GO:1900434 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 1.1 | 4.3 | GO:0000461 | endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000461) |
| 0.9 | 2.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.9 | 2.6 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.7 | 2.8 | GO:0000296 | spermine transport(GO:0000296) |
| 0.7 | 2.8 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.7 | 4.7 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.6 | 6.8 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.6 | 2.4 | GO:0014070 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 0.6 | 2.3 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.5 | 4.1 | GO:0000372 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 0.5 | 2.0 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.5 | 1.4 | GO:0008272 | sulfate transport(GO:0008272) C4-dicarboxylate transport(GO:0015740) |
| 0.4 | 2.6 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.4 | 1.2 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.4 | 1.2 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) L-kynurenine catabolic process(GO:0097053) |
| 0.4 | 2.7 | GO:0015791 | polyol transport(GO:0015791) |
| 0.4 | 1.5 | GO:0090337 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 0.3 | 2.0 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 0.3 | 49.4 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.3 | 2.3 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.3 | 0.9 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.3 | 1.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.3 | 1.2 | GO:0009240 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.3 | 0.9 | GO:0071454 | response to anoxia(GO:0034059) cellular response to anoxia(GO:0071454) |
| 0.3 | 1.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.3 | 1.5 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.2 | 2.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 2.6 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.2 | 1.6 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.2 | 0.6 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 0.2 | 1.1 | GO:0050810 | regulation of steroid metabolic process(GO:0019218) regulation of ergosterol biosynthetic process(GO:0032443) regulation of steroid biosynthetic process(GO:0050810) |
| 0.2 | 0.8 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.2 | 0.9 | GO:0060277 | obsolete negative regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0060277) |
| 0.2 | 0.6 | GO:0019379 | sulfate assimilation, phosphoadenylyl sulfate reduction by phosphoadenylyl-sulfate reductase (thioredoxin)(GO:0019379) sulfate reduction(GO:0019419) |
| 0.2 | 1.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 0.2 | GO:0032233 | regulation of actin filament bundle assembly(GO:0032231) positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.2 | 0.5 | GO:1900062 | regulation of cell aging(GO:0090342) regulation of replicative cell aging(GO:1900062) |
| 0.2 | 0.5 | GO:0030541 | plasmid partitioning(GO:0030541) |
| 0.2 | 0.6 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.2 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.2 | 0.5 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.2 | 0.3 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.6 | GO:0051597 | response to methylmercury(GO:0051597) cellular response to methylmercury(GO:0071406) |
| 0.1 | 0.6 | GO:0015833 | peptide transport(GO:0015833) |
| 0.1 | 1.3 | GO:0055069 | cellular zinc ion homeostasis(GO:0006882) zinc ion homeostasis(GO:0055069) |
| 0.1 | 0.6 | GO:0007157 | agglutination involved in conjugation with cellular fusion(GO:0000752) agglutination involved in conjugation(GO:0000771) heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) cell-cell adhesion(GO:0098609) adhesion between unicellular organisms(GO:0098610) multi organism cell adhesion(GO:0098740) cell-cell adhesion via plasma-membrane adhesion molecules(GO:0098742) |
| 0.1 | 0.3 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.1 | 2.5 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 0.1 | 0.5 | GO:0009097 | isoleucine biosynthetic process(GO:0009097) |
| 0.1 | 0.8 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.1 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.1 | 0.5 | GO:0071041 | antisense RNA transcript catabolic process(GO:0071041) |
| 0.1 | 0.6 | GO:0006336 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-independent nucleosome assembly(GO:0006336) DNA replication-dependent nucleosome organization(GO:0034723) positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 0.4 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.1 | 1.6 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.3 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) maintenance of chromatin silencing at telomere(GO:0035392) |
| 0.1 | 1.1 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.1 | 0.6 | GO:0006768 | biotin metabolic process(GO:0006768) biotin biosynthetic process(GO:0009102) |
| 0.1 | 2.9 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.1 | 2.0 | GO:0006469 | negative regulation of protein kinase activity(GO:0006469) |
| 0.1 | 0.4 | GO:0015809 | arginine transport(GO:0015809) |
| 0.1 | 2.2 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.1 | 0.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 0.8 | GO:0000084 | mitotic S phase(GO:0000084) S phase(GO:0051320) |
| 0.1 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.2 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.1 | 0.7 | GO:0006458 | 'de novo' protein folding(GO:0006458) |
| 0.1 | 0.1 | GO:0043068 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.1 | 0.6 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.5 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.1 | 0.6 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 0.1 | 0.3 | GO:0001676 | long-chain fatty acid metabolic process(GO:0001676) |
| 0.1 | 0.6 | GO:0016479 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) |
| 0.1 | 0.2 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.1 | 0.3 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.1 | 1.4 | GO:0044108 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.1 | 1.5 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.1 | 0.8 | GO:0007004 | RNA-dependent DNA biosynthetic process(GO:0006278) telomere maintenance via telomerase(GO:0007004) |
| 0.1 | 1.0 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) peptidyl-proline modification(GO:0018208) |
| 0.1 | 0.3 | GO:0070941 | eisosome assembly(GO:0070941) |
| 0.1 | 0.5 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.1 | 0.8 | GO:0001100 | negative regulation of exit from mitosis(GO:0001100) |
| 0.1 | 0.4 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.4 | GO:0031506 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.1 | 0.4 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.1 | 0.6 | GO:0006415 | translational termination(GO:0006415) |
| 0.1 | 0.4 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.0 | 0.2 | GO:0015883 | FAD transport(GO:0015883) |
| 0.0 | 0.3 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0006276 | plasmid maintenance(GO:0006276) |
| 0.0 | 0.5 | GO:0034221 | fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.0 | 0.3 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.0 | 0.6 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.4 | GO:0072503 | cellular divalent inorganic cation homeostasis(GO:0072503) divalent inorganic cation homeostasis(GO:0072507) |
| 0.0 | 0.3 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.6 | GO:0017148 | negative regulation of translation(GO:0017148) negative regulation of cellular amide metabolic process(GO:0034249) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 1.0 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 1.0 | GO:0040020 | regulation of meiotic nuclear division(GO:0040020) |
| 0.0 | 0.8 | GO:0030148 | sphingolipid biosynthetic process(GO:0030148) |
| 0.0 | 3.6 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) nuclear transport(GO:0051169) |
| 0.0 | 0.6 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) regulation of cyclin-dependent protein kinase activity(GO:1904029) |
| 0.0 | 0.2 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0034517 | ribophagy(GO:0034517) |
| 0.0 | 0.3 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.5 | GO:0006513 | protein monoubiquitination(GO:0006513) |
| 0.0 | 0.5 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.3 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.0 | 0.2 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0006827 | high-affinity iron ion transmembrane transport(GO:0006827) |
| 0.0 | 0.7 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0000743 | nuclear migration involved in conjugation with cellular fusion(GO:0000743) |
| 0.0 | 0.2 | GO:0007323 | peptide pheromone maturation(GO:0007323) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 1.6 | GO:0007017 | microtubule-based process(GO:0007017) |
| 0.0 | 0.3 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0000741 | karyogamy(GO:0000741) karyogamy involved in conjugation with cellular fusion(GO:0000742) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:0019358 | nicotinate nucleotide biosynthetic process(GO:0019357) nicotinate nucleotide salvage(GO:0019358) pyridine nucleotide salvage(GO:0019365) nicotinate nucleotide metabolic process(GO:0046497) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:2000284 | positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.5 | GO:0046939 | nucleotide phosphorylation(GO:0046939) |
| 0.0 | 0.0 | GO:0031070 | intronic snoRNA processing(GO:0031070) intronic box C/D snoRNA processing(GO:0034965) |
| 0.0 | 0.2 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.3 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0007535 | donor selection(GO:0007535) |
| 0.0 | 0.5 | GO:0042244 | ascospore wall assembly(GO:0030476) spore wall assembly(GO:0042244) spore wall biogenesis(GO:0070590) ascospore wall biogenesis(GO:0070591) fungal-type cell wall assembly(GO:0071940) |
| 0.0 | 0.2 | GO:0000754 | adaptation of signaling pathway by response to pheromone involved in conjugation with cellular fusion(GO:0000754) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.9 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0044091 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.0 | GO:0009452 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 1.7 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0000917 | barrier septum assembly(GO:0000917) cell septum assembly(GO:0090529) |
| 0.0 | 0.2 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.4 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.5 | 38.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 1.2 | GO:0031518 | CBF3 complex(GO:0031518) |
| 0.3 | 18.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.3 | 0.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.3 | 1.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 1.7 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 1.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.2 | 0.9 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 1.1 | GO:0034518 | RNA cap binding complex(GO:0034518) |
| 0.2 | 0.6 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 0.5 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.2 | 2.4 | GO:0005940 | septin ring(GO:0005940) |
| 0.2 | 1.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.2 | 1.2 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.2 | 1.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.2 | 2.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.2 | 0.8 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.6 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 0.1 | 0.4 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.1 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 3.5 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.4 | GO:0070210 | Rpd3L-Expanded complex(GO:0070210) |
| 0.1 | 0.6 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 0.4 | GO:0030428 | cell septum(GO:0030428) |
| 0.1 | 0.3 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.1 | 0.5 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.1 | 0.4 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.5 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.4 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 0.1 | 0.5 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.1 | 1.0 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.1 | 0.9 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.2 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.7 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 8.5 | GO:0005730 | nucleolus(GO:0005730) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) tubulin complex(GO:0045298) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.3 | GO:0090544 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0044697 | HICS complex(GO:0044697) |
| 0.0 | 0.3 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.4 | GO:0016586 | RSC complex(GO:0016586) |
| 0.0 | 0.1 | GO:0034099 | luminal surveillance complex(GO:0034099) |
| 0.0 | 0.6 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
| 0.0 | 0.5 | GO:0000784 | nuclear chromosome, telomeric region(GO:0000784) |
| 0.0 | 2.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 0.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.8 | GO:0043332 | mating projection tip(GO:0043332) |
| 0.0 | 0.1 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.8 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.1 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.0 | 0.1 | GO:0032126 | eisosome(GO:0032126) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 1.5 | GO:0005933 | cellular bud(GO:0005933) |
| 0.0 | 0.1 | GO:0044453 | nuclear outer membrane(GO:0005640) nuclear membrane part(GO:0044453) |
| 0.0 | 0.8 | GO:0000785 | chromatin(GO:0000785) |
| 0.0 | 0.1 | GO:0098797 | plasma membrane protein complex(GO:0098797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 4.7 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 1.1 | 4.4 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 1.0 | 3.0 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.9 | 2.8 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 0.9 | 2.8 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.9 | 2.6 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.8 | 2.5 | GO:0016872 | intramolecular lyase activity(GO:0016872) |
| 0.7 | 2.2 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.7 | 0.7 | GO:0032451 | demethylase activity(GO:0032451) |
| 0.7 | 2.8 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 0.5 | 4.2 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.5 | 2.7 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.4 | 1.8 | GO:1904680 | peptide transmembrane transporter activity(GO:1904680) |
| 0.4 | 2.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.4 | 1.6 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.4 | 1.6 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.4 | 1.2 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.4 | 1.5 | GO:0016854 | ammonia-lyase activity(GO:0016841) racemase and epimerase activity(GO:0016854) |
| 0.4 | 1.5 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.3 | 2.0 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.3 | 4.3 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.3 | 1.3 | GO:0050072 | m7G(5')pppN diphosphatase activity(GO:0050072) |
| 0.3 | 53.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 0.2 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 0.2 | 2.2 | GO:0051213 | dioxygenase activity(GO:0051213) |
| 0.2 | 1.7 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.2 | 3.8 | GO:0004520 | endodeoxyribonuclease activity(GO:0004520) |
| 0.2 | 1.2 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.2 | 2.4 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.2 | 0.5 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.2 | 2.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.2 | 0.8 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.1 | 1.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 0.5 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.1 | 0.3 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.3 | GO:0004805 | trehalose-phosphatase activity(GO:0004805) |
| 0.1 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 1.3 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.1 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.1 | 0.3 | GO:0004000 | adenosine deaminase activity(GO:0004000) tRNA-specific adenosine deaminase activity(GO:0008251) |
| 0.1 | 0.6 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.1 | 2.9 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.1 | 6.3 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.4 | GO:0042973 | glucan endo-1,3-beta-D-glucosidase activity(GO:0042973) |
| 0.1 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.6 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.1 | 0.3 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.1 | 0.6 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.1 | 0.4 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.1 | 0.4 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.1 | 0.3 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.1 | 0.4 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.1 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 2.0 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.7 | GO:0016859 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) cis-trans isomerase activity(GO:0016859) |
| 0.1 | 0.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.1 | 0.3 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.1 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.1 | 0.3 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.1 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.2 | GO:0015230 | FAD transmembrane transporter activity(GO:0015230) |
| 0.0 | 0.6 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.2 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.5 | GO:0019904 | protein domain specific binding(GO:0019904) |
| 0.0 | 0.4 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.4 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.8 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.0 | 1.5 | GO:0000287 | magnesium ion binding(GO:0000287) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.8 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 1.3 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.2 | GO:0051183 | vitamin transporter activity(GO:0051183) |
| 0.0 | 0.1 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.0 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.0 | 2.2 | GO:0008289 | lipid binding(GO:0008289) |
| 0.0 | 0.1 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.5 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0015665 | alcohol transmembrane transporter activity(GO:0015665) |
| 0.0 | 0.1 | GO:0004396 | hexokinase activity(GO:0004396) |
| 0.0 | 0.4 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.3 | GO:0072509 | divalent inorganic cation transmembrane transporter activity(GO:0072509) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0030515 | snoRNA binding(GO:0030515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.1 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 1.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.1 | 0.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 0.2 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | Genes involved in Downstream signaling of activated FGFR |
| 0.6 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.4 | 3.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.3 | 0.7 | REACTOME REGULATION OF MRNA STABILITY BY PROTEINS THAT BIND AU RICH ELEMENTS | Genes involved in Regulation of mRNA Stability by Proteins that Bind AU-rich Elements |
| 0.3 | 4.9 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.2 | 1.7 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 0.7 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 0.2 | 1.3 | REACTOME METABOLISM OF MRNA | Genes involved in Metabolism of mRNA |
| 0.1 | 0.3 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 0.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.1 | 0.2 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.1 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 0.1 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |