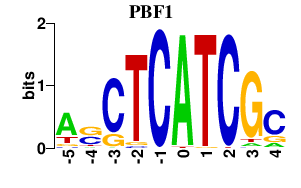
Results for PBF1
Z-value: 2.90
Transcription factors associated with PBF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
TOD6
|
S000000150 | PAC motif binding protein involved in rRNA and ribosome biogenesis |
Activity-expression correlation:
Activity profile of PBF1 motif
Sorted Z-values of PBF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YFR055W | 27.84 |
IRC7
|
Putative cystathionine beta-lyase; involved in copper ion homeostasis and sulfur metabolism; null mutant displays increased levels of spontaneous Rad52p foci; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
|
| YOR101W | 25.80 |
RAS1
|
GTPase involved in G-protein signaling in the adenylate cyclase activating pathway, plays a role in cell proliferation; localized to the plasma membrane; homolog of mammalian RAS proto-oncogenes |
|
| YFR056C | 24.05 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; partially overlaps the uncharacterized gene YFR055W |
||
| YER073W | 21.09 |
ALD5
|
Mitochondrial aldehyde dehydrogenase, involved in regulation or biosynthesis of electron transport chain components and acetate formation; activated by K+; utilizes NADP+ as the preferred coenzyme; constitutively expressed |
|
| YOR096W | 19.06 |
RPS7A
|
Protein component of the small (40S) ribosomal subunit, nearly identical to Rps7Bp; interacts with Kti11p; deletion causes hypersensitivity to zymocin; has similarity to rat S7 and Xenopus S8 ribosomal proteins |
|
| YKR092C | 18.81 |
SRP40
|
Nucleolar, serine-rich protein with a role in preribosome assembly or transport; may function as a chaperone of small nucleolar ribonucleoprotein particles (snoRNPs); immunologically and structurally to rat Nopp140 |
|
| YOR340C | 17.13 |
RPA43
|
RNA polymerase I subunit A43 |
|
| YGR251W | 16.95 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YIR021W | 15.42 |
MRS1
|
Protein required for the splicing of two mitochondrial group I introns (BI3 in COB and AI5beta in COX1); forms a splicing complex, containing four subunits of Mrs1p and two subunits of the BI3-encoded maturase, that binds to the BI3 RNA |
|
| YDL148C | 15.30 |
NOP14
|
Nucleolar protein, forms a complex with Noc4p that mediates maturation and nuclear export of 40S ribosomal subunits; also present in the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YLR196W | 14.74 |
PWP1
|
Protein with WD-40 repeats involved in rRNA processing; associates with trans-acting ribosome biogenesis factors; similar to beta-transducin superfamily |
|
| YLR009W | 14.71 |
RLP24
|
Essential protein with similarity to Rpl24Ap and Rpl24Bp, associated with pre-60S ribosomal subunits and required for ribosomal large subunit biogenesis |
|
| YDL208W | 14.40 |
NHP2
|
Nuclear protein related to mammalian high mobility group (HMG) proteins, essential for function of H/ACA-type snoRNPs, which are involved in 18S rRNA processing |
|
| YML043C | 14.29 |
RRN11
|
Protein required for rDNA transcription by RNA polymerase I, component of the core factor (CF) of rDNA transcription factor, which also contains Rrn6p and Rrn7p |
|
| YPL211W | 13.71 |
NIP7
|
Nucleolar protein required for 60S ribosome subunit biogenesis, constituent of 66S pre-ribosomal particles; physically interacts with Nop8p and the exosome subunit Rrp43p |
|
| YPR148C | 13.63 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm in a punctate pattern |
||
| YLR062C | 13.47 |
BUD28
|
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; 98% of ORF overlaps the verified gene RPL22A; diploid mutant displays a weak budding pattern phenotype in a systematic assay |
|
| YER137C | 13.44 |
Putative protein of unknown function |
||
| YPL143W | 12.86 |
RPL33A
|
N-terminally acetylated ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Bp and has similarity to rat L35a; rpl33a null mutant exhibits slow growth while rpl33a rpl33b double null mutant is inviable |
|
| YGR128C | 12.71 |
UTP8
|
Nucleolar protein required for export of tRNAs from the nucleus; also copurifies with the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YLR355C | 12.47 |
ILV5
|
Acetohydroxyacid reductoisomerase, mitochondrial protein involved in branched-chain amino acid biosynthesis, also required for maintenance of wild-type mitochondrial DNA and found in mitochondrial nucleoids |
|
| YNL112W | 12.24 |
DBP2
|
Essential ATP-dependent RNA helicase of the DEAD-box protein family, involved in nonsense-mediated mRNA decay and rRNA processing |
|
| YHR128W | 12.18 |
FUR1
|
Uracil phosphoribosyltransferase, synthesizes UMP from uracil; involved in the pyrimidine salvage pathway |
|
| YNL248C | 11.94 |
RPA49
|
RNA polymerase I subunit A49 |
|
| YJL010C | 11.73 |
NOP9
|
Essential component of pre-40S ribosomes that is required for early cleavages of 35S pre-rRNA and hence formation of 18S rRNA; binds RNA in vitro and contains multiple pumilio-like repeats |
|
| YML123C | 11.56 |
PHO84
|
High-affinity inorganic phosphate (Pi) transporter and low-affinity manganese transporter; regulated by Pho4p and Spt7p; mutation confers resistance to arsenate; exit from the ER during maturation requires Pho86p |
|
| YDL023C | 11.46 |
Dubious open reading frame, unlikely to encode a protein; not conserved in other Saccharomyces species; overlaps the verified gene GPD1; deletion confers sensitivity to GSAO; deletion in cyr1 mutant results in loss of stress resistance |
||
| YER146W | 11.18 |
LSM5
|
Lsm (Like Sm) protein; part of heteroheptameric complexes (Lsm2p-7p and either Lsm1p or 8p): cytoplasmic Lsm1p complex involved in mRNA decay; nuclear Lsm8p complex part of U6 snRNP and possibly involved in processing tRNA, snoRNA, and rRNA |
|
| YDL051W | 11.16 |
LHP1
|
RNA binding protein required for maturation of tRNA and snRNA precursors; acts as a molecular chaperone for RNAs transcribed by polymerase III; homologous to human La (SS-B) autoantigen |
|
| YDL055C | 11.15 |
PSA1
|
GDP-mannose pyrophosphorylase (mannose-1-phosphate guanyltransferase), synthesizes GDP-mannose from GTP and mannose-1-phosphate in cell wall biosynthesis; required for normal cell wall structure |
|
| YDL022C-A | 11.12 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the verified gene DIA3; identified by fungal homology and RT-PCR |
||
| YIL118W | 11.09 |
RHO3
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins involved in the establishment of cell polarity; GTPase activity positively regulated by the GTPase activating protein (GAP) Rgd1p |
|
| YNL141W | 10.97 |
AAH1
|
Adenine deaminase (adenine aminohydrolase), converts adenine to hypoxanthine; involved in purine salvage; transcriptionally regulated by nutrient levels and growth phase; Aah1p degraded upon entry into quiescence via SCF and the proteasome |
|
| YOR004W | 10.89 |
UTP23
|
Essential nucleolar protein that is a component of the SSU (small subunit) processome involved in 40S ribosomal subunit biogenesis; has homology to PINc domain protein Fcf1p, although the PINc domain of Utp23p is not required for function |
|
| YGR086C | 10.89 |
PIL1
|
Primary component of eisosomes, which are large immobile cell cortex structures associated with endocytosis; null mutants show activation of Pkc1p/Ypk1p stress resistance pathways; detected in phosphorylated state in mitochondria |
|
| YER074W | 10.70 |
RPS24A
|
Protein component of the small (40S) ribosomal subunit; identical to Rps24Bp and has similarity to rat S24 ribosomal protein |
|
| YGR123C | 10.68 |
PPT1
|
Protein serine/threonine phosphatase with similarity to human phosphatase PP5; present in both the nucleus and cytoplasm; expressed during logarithmic growth; computational analyses suggest roles in phosphate metabolism and rRNA processing |
|
| YMR290C | 10.64 |
HAS1
|
ATP-dependent RNA helicase; localizes to both the nuclear periphery and nucleolus; highly enriched in nuclear pore complex fractions; constituent of 66S pre-ribosomal particles |
|
| YLR432W | 10.52 |
IMD3
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YMR290W-A | 10.42 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; overlaps 5' end of essential HAS1 gene which encodes an ATP-dependent RNA helicase |
||
| YNL178W | 10.42 |
RPS3
|
Protein component of the small (40S) ribosomal subunit, has apurinic/apyrimidinic (AP) endonuclease activity; essential for viability; has similarity to E. coli S3 and rat S3 ribosomal proteins |
|
| YJL122W | 10.31 |
ALB1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit; interacts directly with Arx1p; responsible for Tif6p recycling defects in absence of Rei1p |
|
| YBL027W | 10.15 |
RPL19B
|
Protein component of the large (60S) ribosomal subunit, nearly identical to Rpl19Ap and has similarity to rat L19 ribosomal protein; rpl19a and rpl19b single null mutations result in slow growth, while the double null mutation is lethal |
|
| YBR126W-A | 10.13 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR126W-B; identified by gene-trapping, microarray analysis, and genome-wide homology searches |
||
| YBR154C | 10.10 |
RPB5
|
RNA polymerase subunit ABC27, common to RNA polymerases I, II, and III; contacts DNA and affects transactivation |
|
| YBL039C | 10.04 |
URA7
|
Major CTP synthase isozyme (see also URA8), catalyzes the ATP-dependent transfer of the amide nitrogen from glutamine to UTP, forming CTP, the final step in de novo biosynthesis of pyrimidines; involved in phospholipid biosynthesis |
|
| YKL078W | 9.98 |
DHR2
|
Predominantly nucleolar DEAH-box ATP-dependent RNA helicase, required for 18S rRNA synthesis |
|
| YPL012W | 9.98 |
RRP12
|
Protein required for export of the ribosomal subunits; associates with the RNA components of the pre-ribosomes; contains HEAT-repeats |
|
| YNL113W | 9.96 |
RPC19
|
RNA polymerase subunit, common to RNA polymerases I and III |
|
| YCR018C | 9.89 |
SRD1
|
Protein involved in the processing of pre-rRNA to mature rRNA; contains a C2/C2 zinc finger motif; srd1 mutation suppresses defects caused by the rrp1-1 mutation |
|
| YBL028C | 9.81 |
Protein of unknown function that may interact with ribosomes; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus; predicted to be involved in ribosome biogenesis |
||
| YNL110C | 9.71 |
NOP15
|
Constituent of 66S pre-ribosomal particles, involved in 60S ribosomal subunit biogenesis; localizes to both nucleolus and cytoplasm |
|
| YDL022W | 9.63 |
GPD1
|
NAD-dependent glycerol-3-phosphate dehydrogenase, key enzyme of glycerol synthesis, essential for growth under osmotic stress; expression regulated by high-osmolarity glycerol response pathway; homolog of Gpd2p |
|
| YDL014W | 9.56 |
NOP1
|
Nucleolar protein, component of the small subunit processome complex, which is required for processing of pre-18S rRNA; has similarity to mammalian fibrillarin |
|
| YOR153W | 9.50 |
PDR5
|
Plasma membrane ATP-binding cassette (ABC) transporter, short-lived multidrug transporter actively regulated by Pdr1p; also involved in steroid transport, cation resistance, and cellular detoxification during exponential growth |
|
| YOR272W | 9.50 |
YTM1
|
Constituent of 66S pre-ribosomal particles, required for maturation of the large ribosomal subunit |
|
| YCL036W | 9.44 |
GFD2
|
Protein of unknown function, identified as a high-copy suppressor of a dbp5 mutation |
|
| YOL077C | 9.43 |
BRX1
|
Nucleolar protein, constituent of 66S pre-ribosomal particles; depletion leads to defects in rRNA processing and a block in the assembly of large ribosomal subunits; possesses a sigma(70)-like RNA-binding motif |
|
| YKL081W | 9.24 |
TEF4
|
Translation elongation factor EF-1 gamma |
|
| YMR131C | 9.23 |
RRB1
|
Essential nuclear protein involved in early steps of ribosome biogenesis; physically interacts with the ribosomal protein Rpl3p |
|
| YKR081C | 9.21 |
RPF2
|
Essential protein involved in the processing of pre-rRNA and the assembly of the 60S ribosomal subunit; interacts with ribosomal protein L11; localizes predominantly to the nucleolus; constituent of 66S pre-ribosomal particles |
|
| YMR049C | 9.19 |
ERB1
|
Protein required for maturation of the 25S and 5.8S ribosomal RNAs; constituent of 66S pre-ribosomal particles; homologous to mammalian Bop1 |
|
| YMR217W | 9.15 |
GUA1
|
GMP synthase, an enzyme that catalyzes the second step in the biosynthesis of GMP from inosine 5'-phosphate (IMP); transcription is not subject to regulation by guanine but is negatively regulated by nutrient starvation |
|
| YPR010C | 9.08 |
RPA135
|
RNA polymerase I subunit A135 |
|
| YKL009W | 9.06 |
MRT4
|
Protein involved in mRNA turnover and ribosome assembly, localizes to the nucleolus |
|
| YGR151C | 9.05 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF RSR1/BUD1/YGR152C |
||
| YJR070C | 9.03 |
LIA1
|
Deoxyhypusine hydroxylase, a HEAT-repeat containing metalloenzyme that catalyses hypusine formation; binds to and is required for the modification of Hyp2p (eIF5A); complements S. pombe mmd1 mutants defective in mitochondrial positioning |
|
| YDR465C | 8.94 |
RMT2
|
Arginine methyltransferase; ribosomal protein L12 is a substrate |
|
| YML121W | 8.94 |
GTR1
|
Cytoplasmic GTP binding protein and negative regulator of the Ran/Tc4 GTPase cycle; component of GSE complex, which is required for sorting of Gap1p; involved in phosphate transport and telomeric silencing; similar to human RagA and RagB |
|
| YHR066W | 8.81 |
SSF1
|
Constituent of 66S pre-ribosomal particles, required for ribosomal large subunit maturation; functionally redundant with Ssf2p; member of the Brix family |
|
| YPL212C | 8.79 |
PUS1
|
tRNA:pseudouridine synthase, introduces pseudouridines at positions 26-28, 34-36, 65, and 67 of tRNA; nuclear protein that appears to be involved in tRNA export; also acts on U2 snRNA |
|
| YNL175C | 8.77 |
NOP13
|
Protein of unknown function, localizes to the nucleolus and nucleoplasm; contains an RNA recognition motif (RRM) and has similarity to Nop12p, which is required for processing of pre-18S rRNA |
|
| YLR075W | 8.68 |
RPL10
|
Protein component of the large (60S) ribosomal subunit, responsible for joining the 40S and 60S subunits; regulates translation initiation; has similarity to rat L10 ribosomal protein and to members of the QM gene family |
|
| YOL086C | 8.60 |
ADH1
|
Alcohol dehydrogenase, fermentative isozyme active as homo- or heterotetramers; required for the reduction of acetaldehyde to ethanol, the last step in the glycolytic pathway |
|
| YOL127W | 8.56 |
RPL25
|
Primary rRNA-binding ribosomal protein component of the large (60S) ribosomal subunit, has similarity to E. coli L23 and rat L23a ribosomal proteins; binds to 26S rRNA via a conserved C-terminal motif |
|
| YLR413W | 8.54 |
Putative protein of unknown function; YLR413W is not an essential gene |
||
| YJR071W | 8.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YEL053W-A | 8.48 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified gene YEL054C |
||
| YKL164C | 8.39 |
PIR1
|
O-glycosylated protein required for cell wall stability; attached to the cell wall via beta-1,3-glucan; mediates mitochondrial translocation of Apn1p; expression regulated by the cell integrity pathway and by Swi5p during the cell cycle |
|
| YFR001W | 8.38 |
LOC1
|
Nuclear protein involved in asymmetric localization of ASH1 mRNA; binds double-stranded RNA in vitro; constituent of 66S pre-ribosomal particles |
|
| YGR124W | 8.35 |
ASN2
|
Asparagine synthetase, isozyme of Asn1p; catalyzes the synthesis of L-asparagine from L-aspartate in the asparagine biosynthetic pathway |
|
| YOR310C | 8.33 |
NOP58
|
Protein involved in pre-rRNA processing, 18S rRNA synthesis, and snoRNA synthesis; component of the small subunit processome complex, which is required for processing of pre-18S rRNA |
|
| YIR026C | 8.33 |
YVH1
|
Protein phosphatase involved in vegetative growth at low temperatures, sporulation, and glycogen accumulation; transcription induced by low temperature and nitrogen starvation; member of the dual-specificity family of protein phosphatases |
|
| YLR068W | 8.27 |
FYV7
|
Protein of unknown function, required for survival upon exposure to K1 killer toxin; involved in processing the 35S rRNA primary transcript to generate the 20S and 27SA2 pre-rRNA transcripts |
|
| YNR053C | 8.26 |
NOG2
|
Putative GTPase that associates with pre-60S ribosomal subunits in the nucleolus and is required for their nuclear export and maturation |
|
| YAL059W | 8.24 |
ECM1
|
Protein of unknown function, localized in the nucleoplasm and the nucleolus, genetically interacts with MTR2 in 60S ribosomal protein subunit export |
|
| YOR226C | 8.23 |
ISU2
|
Conserved protein of the mitochondrial matrix, required for synthesis of mitochondrial and cytosolic iron-sulfur proteins, performs a scaffolding function in mitochondria during Fe/S cluster assembly; isu1 isu2 double mutant is inviable |
|
| YDR087C | 8.19 |
RRP1
|
Essential evolutionarily conserved nucleolar protein necessary for biogenesis of 60S ribosomal subunits and processing of pre-rRNAs to mature rRNAs, associated with several distinct 66S pre-ribosomal particles |
|
| YNL089C | 8.08 |
Dubious open reading frame unlikely to encode a functional protein; almost completely overlaps YNL090W/RHO2 which encodes a small GTPase of the Rho/Rac subfamily of Ras-like proteins |
||
| YGL029W | 8.01 |
CGR1
|
Protein involved in nucleolar integrity and processing of the pre-rRNA for the 60S ribosome subunit; transcript is induced in response to cytotoxic stress but not genotoxic stress |
|
| YLR180W | 7.97 |
SAM1
|
S-adenosylmethionine synthetase, catalyzes transfer of the adenosyl group of ATP to the sulfur atom of methionine; one of two differentially regulated isozymes (Sam1p and Sam2p) |
|
| YDR399W | 7.95 |
HPT1
|
Dimeric hypoxanthine-guanine phosphoribosyltransferase, catalyzes the formation of both inosine monophosphate and guanosine monophosphate; mutations in the human homolog HPRT1 can cause Lesch-Nyhan syndrome and Kelley-Seegmiller syndrome |
|
| YGR145W | 7.93 |
ENP2
|
Essential nucleolar protein of unknown function; contains WD repeats, interacts with Mpp10p and Bfr2p, and has homology to Spb1p |
|
| YJL148W | 7.89 |
RPA34
|
RNA polymerase I subunit A34.5 |
|
| YGR159C | 7.87 |
NSR1
|
Nucleolar protein that binds nuclear localization sequences, required for pre-rRNA processing and ribosome biogenesis |
|
| YGL055W | 7.85 |
OLE1
|
Delta(9) fatty acid desaturase, required for monounsaturated fatty acid synthesis and for normal distribution of mitochondria |
|
| YPL093W | 7.82 |
NOG1
|
Putative GTPase that associates with free 60S ribosomal subunits in the nucleolus and is required for 60S ribosomal subunit biogenesis; constituent of 66S pre-ribosomal particles; member of the ODN family of nucleolar G-proteins |
|
| YOR108W | 7.76 |
LEU9
|
Alpha-isopropylmalate synthase II (2-isopropylmalate synthase), catalyzes the first step in the leucine biosynthesis pathway; the minor isozyme, responsible for the residual alpha-IPMS activity detected in a leu4 null mutant |
|
| YKL172W | 7.76 |
EBP2
|
Essential protein required for the maturation of 25S rRNA and 60S ribosomal subunit assembly, localizes to the nucleolus; constituent of 66S pre-ribosomal particles |
|
| YDR398W | 7.74 |
UTP5
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YLR154C | 7.69 |
RNH203
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YIL096C | 7.65 |
Putative protein of unknown function; associates with precursors of the 60S ribosomal subunit |
||
| YOR271C | 7.59 |
FSF1
|
Putative protein, predicted to be an alpha-isopropylmalate carrier; belongs to the sideroblastic-associated protein family; non-tagged protein is detected in purified mitochondria; likely to play a role in iron homeostasis |
|
| YDL150W | 7.58 |
RPC53
|
RNA polymerase III subunit C53 |
|
| YOL041C | 7.56 |
NOP12
|
Nucleolar protein, required for pre-25S rRNA processing; contains an RNA recognition motif (RRM) and has similarity to Nop13p, Nsr1p, and putative orthologs in Drosophila and S. pombe |
|
| YGL078C | 7.53 |
DBP3
|
Putative ATP-dependent RNA helicase of the DEAD-box family involved in ribosomal biogenesis |
|
| YFL015W-A | 7.52 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YBR249C | 7.52 |
ARO4
|
3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, catalyzes the first step in aromatic amino acid biosynthesis and is feedback-inhibited by tyrosine or high concentrations of phenylalanine or tryptophan |
|
| YHR148W | 7.51 |
IMP3
|
Component of the SSU processome, which is required for pre-18S rRNA processing, essential protein that interacts with Mpp10p and mediates interactions of Imp4p and Mpp10p with U3 snoRNA |
|
| YGR280C | 7.49 |
PXR1
|
Essential protein involved in rRNA and snoRNA maturation; competes with TLC1 RNA for binding to Est2p, suggesting a role in negative regulation of telomerase; human homolog inhibits telomerase; contains a G-patch RNA interacting domain |
|
| YIL091C | 7.45 |
Protein required for cell viability |
||
| YER102W | 7.42 |
RPS8B
|
Protein component of the small (40S) ribosomal subunit; identical to Rps8Ap and has similarity to rat S8 ribosomal protein |
|
| YOR342C | 7.38 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and the nucleus |
||
| YPL108W | 7.38 |
Cytoplasmic protein of unknown function; non-essential gene that is induced in a GDH1 deleted strain with altered redox metabolism; GFP-fusion protein is induced in response to the DNA-damaging agent MMS |
||
| YDR385W | 7.32 |
EFT2
|
Elongation factor 2 (EF-2), also encoded by EFT1; catalyzes ribosomal translocation during protein synthesis; contains diphthamide, the unique posttranslationally modified histidine residue specifically ADP-ribosylated by diphtheria toxin |
|
| YOR234C | 7.26 |
RPL33B
|
Ribosomal protein L37 of the large (60S) ribosomal subunit, nearly identical to Rpl33Ap and has similarity to rat L35a; rpl33b null mutant exhibits normal growth while rpl33a rpl33b double null mutant is inviable |
|
| YFL015C | 7.25 |
Dubious open reading frame unlikely to encode a protein; partially overlaps dubious ORF YFL015W-A; YFL015C is not an essential gene |
||
| YGR103W | 7.23 |
NOP7
|
Nucleolar protein involved in rRNA processing and 60S ribosomal subunit biogenesis; constituent of several different pre-ribosomal particles; required for exit from G0 and the initiation of cell proliferation |
|
| YOL124C | 7.21 |
TRM11
|
Catalytic subunit of an adoMet-dependent tRNA methyltransferase complex (Trm11p-Trm112p), required for the methylation of the guanosine nucleotide at position 10 (m2G10) in tRNAs; contains a THUMP domain and a methyltransferase domain |
|
| YPL037C | 7.09 |
EGD1
|
Subunit beta1 of the nascent polypeptide-associated complex (NAC) involved in protein targeting, associated with cytoplasmic ribosomes; enhances DNA binding of the Gal4p activator; homolog of human BTF3b |
|
| YKR024C | 7.03 |
DBP7
|
Putative ATP-dependent RNA helicase of the DEAD-box family involved in ribosomal biogenesis |
|
| YCL059C | 7.00 |
KRR1
|
Essential nucleolar protein required for the synthesis of 18S rRNA and for the assembly of 40S ribosomal subunit |
|
| YGL120C | 7.00 |
PRP43
|
RNA helicase in the DEAH-box family, functions in both RNA polymerase I and polymerase II transcript metabolism, involved in release of the lariat-intron from the spliceosome |
|
| YOL080C | 6.99 |
REX4
|
Putative RNA exonuclease possibly involved in pre-rRNA processing and ribosome assembly |
|
| YDR060W | 6.96 |
MAK21
|
Constituent of 66S pre-ribosomal particles, required for large (60S) ribosomal subunit biogenesis; involved in nuclear export of pre-ribosomes; required for maintenance of dsRNA virus; homolog of human CAATT-binding protein |
|
| YBL092W | 6.92 |
RPL32
|
Protein component of the large (60S) ribosomal subunit, has similarity to rat L32 ribosomal protein; overexpression disrupts telomeric silencing |
|
| YJL200C | 6.92 |
ACO2
|
Putative mitochondrial aconitase isozyme; similarity to Aco1p, an aconitase required for the TCA cycle; expression induced during growth on glucose, by amino acid starvation via Gcn4p, and repressed on ethanol |
|
| YBR126C | 6.91 |
TPS1
|
Synthase subunit of trehalose-6-phosphate synthase/phosphatase complex, which synthesizes the storage carbohydrate trehalose; also found in a monomeric form; expression is induced by the stress response and repressed by the Ras-cAMP pathway |
|
| YDL112W | 6.88 |
TRM3
|
2'-O-ribose methyltransferase, catalyzes the ribose methylation of the guanosine nucleotide at position 18 of tRNAs |
|
| YBR142W | 6.88 |
MAK5
|
Essential nucleolar protein, putative DEAD-box RNA helicase required for maintenance of M1 dsRNA virus; involved in biogenesis of large (60S) ribosomal subunits |
|
| YLR409C | 6.87 |
UTP21
|
Possible U3 snoRNP protein involved in maturation of pre-18S rRNA, based on computational analysis of large-scale protein-protein interaction data |
|
| YLR276C | 6.87 |
DBP9
|
ATP-dependent RNA helicase of the DEAD-box family involved in biogenesis of the 60S ribosomal subunit |
|
| YAL025C | 6.85 |
MAK16
|
Essential nuclear protein, constituent of 66S pre-ribosomal particles; required for maturation of 25S and 5.8S rRNAs; required for maintenance of M1 satellite double-stranded RNA of the L-A virus |
|
| YGR245C | 6.82 |
SDA1
|
Highly conserved nuclear protein required for actin cytoskeleton organization and passage through Start, plays a critical role in G1 events, binds Nap1p, also involved in 60S ribosome biogenesis |
|
| YOL079W | 6.76 |
Dubious open reading frame, unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YDL062W | 6.74 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps uncharacterized ORF YDL063C; YDL062W is not essential |
||
| YHR089C | 6.73 |
GAR1
|
Protein component of the H/ACA snoRNP pseudouridylase complex, involved in the modification and cleavage of the 18S pre-rRNA |
|
| YER126C | 6.71 |
NSA2
|
Protein constituent of 66S pre-ribosomal particles, contributes to processing of the 27S pre-rRNA |
|
| YMR123W | 6.71 |
PKR1
|
V-ATPase assembly factor, functions with other V-ATPase assembly factors in the ER to efficiently assemble the V-ATPase membrane sector (V0); overproduction confers resistance to Pichia farinosa killer toxin |
|
| YJL050W | 6.68 |
MTR4
|
Dead-box family ATP dependent helicase required for mRNA export from the nucleus; co-factor of the exosome complex, required for 3' end formation of 5.8S rRNA |
|
| YDR279W | 6.65 |
RNH202
|
Ribonuclease H2 subunit, required for RNase H2 activity |
|
| YFR054C | 6.65 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDR120C | 6.63 |
TRM1
|
tRNA methyltransferase, localizes to both the nucleus and mitochondrion to produce the modified base N2,N2-dimethylguanosine in tRNAs in both compartments |
|
| YLR154W-A | 6.62 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YKL099C | 6.62 |
UTP11
|
Nucleolar protein, component of the small subunit (SSU) processome containing the U3 snoRNA that is involved in processing of pre-18S rRNA |
|
| YER001W | 6.62 |
MNN1
|
Alpha-1,3-mannosyltransferase, integral membrane glycoprotein of the Golgi complex, required for addition of alpha1,3-mannose linkages to N-linked and O-linked oligosaccharides, one of five S. cerevisiae proteins of the MNN1 family |
|
| YKL096W-A | 6.62 |
CWP2
|
Covalently linked cell wall mannoprotein, major constituent of the cell wall; plays a role in stabilizing the cell wall; involved in low pH resistance; precursor is GPI-anchored |
|
| YKR093W | 6.62 |
PTR2
|
Integral membrane peptide transporter, mediates transport of di- and tri-peptides; conserved protein that contains 12 transmembrane domains; PTR2 expression is regulated by the N-end rule pathway via repression by Cup9p |
|
| YNL247W | 6.56 |
Essential protein of unknown function; may interact with ribosomes, based on co-purification experiments |
||
| YLR154W-B | 6.56 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YML056C | 6.55 |
IMD4
|
Inosine monophosphate dehydrogenase, catalyzes the first step of GMP biosynthesis, member of a four-gene family in S. cerevisiae, constitutively expressed |
|
| YKR043C | 6.51 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus |
||
| YNL114C | 6.49 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the verified ORF RPC19/YNL113W, an RNA polymerase subunit |
||
| YLL008W | 6.46 |
DRS1
|
Nucleolar DEAD-box protein required for ribosome assembly and function, including synthesis of 60S ribosomal subunits; constituent of 66S pre-ribosomal particles |
|
| YDL063C | 6.40 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and nucleus; YDL063C is not an essential gene |
||
| YKL014C | 6.37 |
URB1
|
Nucleolar protein required for the normal accumulation of 25S and 5.8S rRNAs, associated with the 27SA2 pre-ribosomal particle; proposed to be involved in the biogenesis of the 60S ribosomal subunit |
|
| YNR003C | 6.37 |
RPC34
|
RNA polymerase III subunit C34; interacts with TFIIIB70 and is a key determinant in pol III recruitment by the preinitiation complex |
|
| YLR129W | 6.36 |
DIP2
|
Nucleolar protein, specifically associated with the U3 snoRNA, part of the large ribonucleoprotein complex known as the small subunit (SSU) processome, required for 18S rRNA biogenesis, part of the active pre-rRNA processing complex |
|
| YNL002C | 6.35 |
RLP7
|
Nucleolar protein with similarity to large ribosomal subunit L7 proteins; constituent of 66S pre-ribosomal particles; plays an essential role in processing of precursors to the large ribosomal subunit RNAs |
|
| YPL086C | 6.34 |
ELP3
|
Subunit of Elongator complex, which is required for modification of wobble nucleosides in tRNA; exhibits histone acetyltransferase activity that is directed to histones H3 and H4; disruption confers resistance to K. lactis zymotoxin |
|
| YGR155W | 6.34 |
CYS4
|
Cystathionine beta-synthase, catalyzes the synthesis of cystathionine from serine and homocysteine, the first committed step in cysteine biosynthesis |
|
| YJL009W | 6.29 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps CCT8/YJL008C, a verified gene encoding a subunit of the cytosolic chaperonin Cct ring complex |
||
| YDL153C | 6.28 |
SAS10
|
Component of the small (ribosomal) subunit (SSU) processosome required for pre-18S rRNa processing; essential nucleolar protein that, when overproduced, disrupts silencing |
|
| YOR243C | 6.27 |
PUS7
|
Pseudouridine synthase, catalyzes pseudouridylation at position 35 in U2 snRNA, position 13 in cytoplasmic tRNAs, and position 35 in pre-tRNA(Tyr); Asp(256) mutation abolishes activity; conserved in archaea, some bacteria, and vertebrates |
|
| YLL011W | 6.23 |
SOF1
|
Essential protein required for biogenesis of 40S (small) ribosomal subunit; has similarity to the beta subunit of trimeric G-proteins and the splicing factor Prp4p |
|
| YBR244W | 6.22 |
GPX2
|
Phospholipid hydroperoxide glutathione peroxidase induced by glucose starvation that protects cells from phospholipid hydroperoxides and nonphospholipid peroxides during oxidative stress |
|
| YHR070C-A | 6.21 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene TRM5/YHR070W |
||
| YNL130C | 6.21 |
CPT1
|
Cholinephosphotransferase, required for phosphatidylcholine biosynthesis and for inositol-dependent regulation of EPT1 transcription |
|
| YCL063W | 6.20 |
VAC17
|
Protein involved in vacuole inheritance; acts as a vacuole-specific receptor for myosin Myo2p |
|
| YHR052W | 6.19 |
CIC1
|
Essential protein that interacts with proteasome components and has a potential role in proteasome substrate specificity; also copurifies with 66S pre-ribosomal particles |
|
| YIL056W | 6.17 |
VHR1
|
Transcriptional activator, required for the vitamin H-responsive element (VHRE) mediated induction of VHT1 (Vitamin H transporter) and BIO5 (biotin biosynthesis intermediate transporter) in response to low biotin concentrations |
|
| YPR163C | 6.16 |
TIF3
|
Translation initiation factor eIF-4B, has RNA annealing activity; contains an RNA recognition motif and binds to single-stranded RNA |
|
| YOR206W | 6.12 |
NOC2
|
Protein that forms a nucleolar complex with Mak21p that binds to 90S and 66S pre-ribosomes, as well as a nuclear complex with Noc3p that binds to 66S pre-ribosomes; both complexes mediate intranuclear transport of ribosomal precursors |
|
| YJR097W | 6.11 |
JJJ3
|
Protein of unknown function, contains a J-domain, which is a region with homology to the E. coli DnaJ protein |
|
| YLR363W-A | 6.04 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YPL239W | 6.03 |
YAR1
|
Cytoplasmic ankyrin-repeat containing protein of unknown function, proposed to link the processes of 40S ribosomal subunit biogenesis and adaptation to osmotic and oxidative stress; expression repressed by heat shock |
|
| YOR341W | 6.02 |
RPA190
|
RNA polymerase I subunit; largest subunit of RNA polymerase I |
|
| YDR101C | 6.02 |
ARX1
|
Shuttling pre-60S factor; involved in the biogenesis of ribosomal large subunit biogenesis; interacts directly with Alb1; responsible for Tif6 recycling defects in absence of Rei1; associated with the ribosomal export complex |
|
| YEL001C | 6.02 |
IRC22
|
Putative protein of unknown function; green fluorescent protein (GFP)-fusion localizes to the ER; YEL001C is non-essential; null mutant displays increased levels of spontaneous Rad52p foci |
|
| YHR197W | 5.99 |
RIX1
|
Essential component of the Rix1 complex (Rix1p, Ipi1p, Ipi3p) that is required for processing of ITS2 sequences from 35S pre-rRNA; Rix1 complex associates with Mdn1p in pre-60S ribosomal particles |
|
| YDR450W | 5.97 |
RPS18A
|
Protein component of the small (40S) ribosomal subunit; nearly identical to Rps18Bp and has similarity to E. coli S13 and rat S18 ribosomal proteins |
|
| YER049W | 5.96 |
TPA1
|
Protein of unknown function; interacts with Sup45p (eRF1), Sup35p (eRF3) and Pab1p; has a role in translation termination efficiency, mRNA poly(A) tail length and mRNA stability |
|
| YOR026W | 5.94 |
BUB3
|
Kinetochore checkpoint WD40 repeat protein that localizes to kinetochores during prophase and metaphase, delays anaphase in the presence of unattached kinetochores; forms complexes with Mad1p-Bub1p and with Cdc20p, binds Mad2p and Mad3p |
|
| YPR074C | 5.88 |
TKL1
|
Transketolase, similar to Tkl2p; catalyzes conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway; needed for synthesis of aromatic amino acids |
|
| YDR021W | 5.88 |
FAL1
|
Nucleolar protein required for maturation of 18S rRNA, member of the eIF4A subfamily of DEAD-box ATP-dependent RNA helicases |
|
| YKL218C | 5.87 |
SRY1
|
3-hydroxyaspartate dehydratase, deaminates L-threo-3-hydroxyaspartate to form oxaloacetate and ammonia; required for survival in the presence of hydroxyaspartate |
|
| YHR169W | 5.79 |
DBP8
|
Putative ATP-dependent RNA helicase of the DEAD-box family involved in biogenesis of the 40S ribosomal subunit |
|
| YDR361C | 5.75 |
BCP1
|
Essential protein involved in nuclear export of Mss4p, which is a lipid kinase that generates phosphatidylinositol 4,5-biphosphate and plays a role in actin cytoskeleton organization and vesicular transport |
|
| YDR083W | 5.74 |
RRP8
|
Nucleolar protein involved in rRNA processing, pre-rRNA cleavage at site A2; also involved in telomere maintenance; mutation is synthetically lethal with a gar1 mutation |
|
| YGR200C | 5.73 |
ELP2
|
Subunit of Elongator complex, which is required for modification of wobble nucleosides in tRNA; target of Kluyveromyces lactis zymocin |
|
| YFR039C | 5.70 |
Putative protein of unknown function; may be involved in response to high salt and changes in carbon source |
||
| YBL070C | 5.67 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YOL076W | 5.65 |
MDM20
|
Non-catalytic subunit of the NatB N-terminal acetyltransferase, which catalyzes N-acetylation of proteins with specific N-terminal sequences; involved in mitochondrial inheritance and actin assembly |
|
| YOR369C | 5.63 |
RPS12
|
Protein component of the small (40S) ribosomal subunit; has similarity to rat ribosomal protein S12 |
|
| YCR016W | 5.62 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus and nucleus; YCR016W is not an essential gene |
||
| YHR144C | 5.60 |
DCD1
|
Deoxycytidine monophosphate (dCMP) deaminase required for dCTP and dTTP synthesis; expression is NOT cell cycle regulated |
|
| YGL171W | 5.60 |
ROK1
|
ATP-dependent RNA helicase of the DEAD box family; required for 18S rRNA synthesis |
|
| YHR149C | 5.59 |
SKG6
|
Integral membrane protein that localizes primarily to growing sites such as the bud tip or the cell periphery; potential Cdc28p substrate; Skg6p interacts with Zds1p and Zds2p |
|
| YGR271C-A | 5.58 |
EFG1
|
Protein of unknown function; null mutant is sensitive to hydroxyurea and is delayed in recovering from alpha-factor arrest; green fluorescent protein (GFP)-fusion protein localizes to the nucleolus |
|
| YPL266W | 5.58 |
DIM1
|
Essential 18S rRNA dimethylase (dimethyladenosine transferase), responsible for conserved m6(2)Am6(2)A dimethylation in 3'-terminal loop of 18S rRNA, part of 90S and 40S pre-particles in nucleolus, involved in pre-ribosomal RNA processing |
Network of associatons between targets according to the STRING database.
First level regulatory network of PBF1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.4 | 28.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 8.6 | 25.8 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 6.8 | 34.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 6.6 | 19.9 | GO:0034310 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 6.1 | 24.3 | GO:0031118 | rRNA pseudouridine synthesis(GO:0031118) |
| 5.5 | 5.5 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 5.3 | 21.2 | GO:0006361 | transcription initiation from RNA polymerase I promoter(GO:0006361) |
| 5.1 | 15.2 | GO:0040031 | snRNA pseudouridine synthesis(GO:0031120) snRNA modification(GO:0040031) |
| 4.8 | 14.3 | GO:0046083 | purine nucleobase salvage(GO:0043096) adenine metabolic process(GO:0046083) |
| 4.7 | 132.4 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 4.5 | 9.0 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 4.5 | 13.5 | GO:0018216 | peptidyl-arginine modification(GO:0018195) peptidyl-arginine methylation(GO:0018216) |
| 4.3 | 12.8 | GO:0034471 | rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 4.0 | 11.9 | GO:0071049 | nuclear retention of pre-mRNA with aberrant 3'-ends at the site of transcription(GO:0071049) |
| 4.0 | 312.7 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 3.9 | 55.3 | GO:0042790 | transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:0042790) |
| 3.8 | 15.1 | GO:0032262 | pyrimidine ribonucleotide salvage(GO:0010138) pyrimidine nucleotide salvage(GO:0032262) pyrimidine nucleoside salvage(GO:0043097) UMP salvage(GO:0044206) |
| 3.7 | 11.2 | GO:0019673 | GDP-mannose biosynthetic process(GO:0009298) GDP-mannose metabolic process(GO:0019673) |
| 3.5 | 17.6 | GO:0031167 | rRNA methylation(GO:0031167) |
| 3.4 | 3.4 | GO:0032785 | negative regulation of DNA-templated transcription, elongation(GO:0032785) |
| 3.4 | 10.1 | GO:0009208 | CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) |
| 3.0 | 54.6 | GO:0030488 | tRNA methylation(GO:0030488) |
| 3.0 | 53.7 | GO:0009304 | tRNA transcription(GO:0009304) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 2.8 | 14.2 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 2.8 | 8.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 2.8 | 11.1 | GO:0090337 | regulation of formin-nucleated actin cable assembly(GO:0090337) positive regulation of formin-nucleated actin cable assembly(GO:0090338) |
| 2.8 | 24.8 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 2.7 | 59.3 | GO:0033750 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 2.7 | 29.5 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 2.6 | 7.8 | GO:0033559 | unsaturated fatty acid biosynthetic process(GO:0006636) unsaturated fatty acid metabolic process(GO:0033559) |
| 2.6 | 143.0 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 2.6 | 7.7 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 2.5 | 219.1 | GO:0006364 | rRNA processing(GO:0006364) |
| 2.4 | 9.8 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 2.4 | 7.3 | GO:0045901 | positive regulation of translational elongation(GO:0045901) |
| 2.4 | 9.6 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 2.4 | 2.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 2.4 | 9.5 | GO:0046898 | response to organic cyclic compound(GO:0014070) response to cycloheximide(GO:0046898) |
| 2.3 | 11.5 | GO:0009098 | leucine biosynthetic process(GO:0009098) |
| 2.2 | 6.6 | GO:0009221 | deoxyribonucleoside monophosphate biosynthetic process(GO:0009157) deoxyribonucleoside monophosphate metabolic process(GO:0009162) pyrimidine deoxyribonucleoside monophosphate metabolic process(GO:0009176) pyrimidine deoxyribonucleoside monophosphate biosynthetic process(GO:0009177) pyrimidine deoxyribonucleotide metabolic process(GO:0009219) pyrimidine deoxyribonucleotide biosynthetic process(GO:0009221) 2'-deoxyribonucleotide biosynthetic process(GO:0009265) 2'-deoxyribonucleotide metabolic process(GO:0009394) deoxyribose phosphate biosynthetic process(GO:0046385) |
| 2.2 | 13.2 | GO:0009099 | valine biosynthetic process(GO:0009099) |
| 2.1 | 12.4 | GO:0070941 | eisosome assembly(GO:0070941) |
| 2.0 | 14.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 2.0 | 8.2 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 2.0 | 8.1 | GO:0000296 | spermine transport(GO:0000296) |
| 2.0 | 8.0 | GO:0046500 | S-adenosylmethionine metabolic process(GO:0046500) |
| 2.0 | 15.8 | GO:0000376 | Group I intron splicing(GO:0000372) RNA splicing, via transesterification reactions with guanosine as nucleophile(GO:0000376) |
| 1.9 | 9.6 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 1.9 | 1.9 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 1.9 | 7.5 | GO:0009423 | chorismate biosynthetic process(GO:0009423) |
| 1.9 | 9.3 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 1.8 | 5.5 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 1.8 | 5.4 | GO:0046901 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 1.8 | 5.4 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) peptidyl-methionine modification(GO:0018206) |
| 1.8 | 5.4 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 1.7 | 5.2 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 1.7 | 5.0 | GO:0006114 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 1.7 | 5.0 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 1.6 | 1.6 | GO:0032447 | protein urmylation(GO:0032447) |
| 1.6 | 6.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 1.5 | 8.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 1.5 | 7.3 | GO:0070814 | hydrogen sulfide metabolic process(GO:0070813) hydrogen sulfide biosynthetic process(GO:0070814) |
| 1.4 | 5.4 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 1.2 | 7.5 | GO:0070525 | tRNA threonylcarbamoyladenosine metabolic process(GO:0070525) |
| 1.2 | 8.7 | GO:0019660 | glucose catabolic process(GO:0006007) glycolytic fermentation to ethanol(GO:0019655) glycolytic fermentation(GO:0019660) hexose catabolic process to ethanol(GO:1902707) |
| 1.2 | 4.9 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 1.2 | 4.9 | GO:1900436 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 1.2 | 1.2 | GO:0016077 | snRNA catabolic process(GO:0016076) snoRNA catabolic process(GO:0016077) nuclear polyadenylation-dependent snoRNA catabolic process(GO:0071036) nuclear polyadenylation-dependent snRNA catabolic process(GO:0071037) |
| 1.2 | 4.7 | GO:0035337 | long-chain fatty acid transport(GO:0015909) fatty-acyl-CoA metabolic process(GO:0035337) |
| 1.1 | 113.9 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 1.1 | 8.9 | GO:0042274 | ribosomal small subunit biogenesis(GO:0042274) |
| 1.1 | 23.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) |
| 1.1 | 7.6 | GO:0031365 | N-terminal protein amino acid modification(GO:0031365) |
| 1.1 | 6.5 | GO:0035437 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 1.1 | 5.3 | GO:0000162 | tryptophan biosynthetic process(GO:0000162) indole-containing compound biosynthetic process(GO:0042435) indolalkylamine biosynthetic process(GO:0046219) |
| 1.0 | 10.4 | GO:0006415 | translational termination(GO:0006415) |
| 1.0 | 3.1 | GO:0042256 | mature ribosome assembly(GO:0042256) |
| 1.0 | 4.0 | GO:0031565 | obsolete cytokinesis checkpoint(GO:0031565) |
| 0.9 | 2.8 | GO:0008272 | sulfate transport(GO:0008272) C4-dicarboxylate transport(GO:0015740) |
| 0.9 | 3.7 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.9 | 1.8 | GO:0031684 | heterotrimeric G-protein complex cycle(GO:0031684) |
| 0.9 | 3.7 | GO:0010688 | negative regulation of ribosomal protein gene transcription from RNA polymerase II promoter(GO:0010688) |
| 0.9 | 0.9 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) isoleucine biosynthetic process(GO:0009097) |
| 0.9 | 13.4 | GO:0006446 | regulation of translational initiation(GO:0006446) |
| 0.9 | 3.5 | GO:0048279 | vesicle fusion with endoplasmic reticulum(GO:0048279) |
| 0.8 | 4.1 | GO:0034503 | protein localization to nucleolar rDNA repeats(GO:0034503) |
| 0.8 | 3.3 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.8 | 7.9 | GO:0051231 | mitotic spindle elongation(GO:0000022) spindle elongation(GO:0051231) |
| 0.8 | 3.8 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.7 | 1.5 | GO:0071044 | histone mRNA metabolic process(GO:0008334) histone mRNA catabolic process(GO:0071044) |
| 0.7 | 4.9 | GO:0097549 | heterochromatin organization involved in chromatin silencing(GO:0070868) chromatin organization involved in negative regulation of transcription(GO:0097549) |
| 0.7 | 6.3 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.7 | 2.7 | GO:0030030 | cell projection organization(GO:0030030) cell projection assembly(GO:0030031) |
| 0.7 | 1.4 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.7 | 4.7 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.7 | 2.7 | GO:0042710 | biofilm formation(GO:0042710) |
| 0.7 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.7 | 2.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.6 | 5.8 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.6 | 19.3 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.6 | 2.5 | GO:0009757 | cellular glucose homeostasis(GO:0001678) carbohydrate mediated signaling(GO:0009756) hexose mediated signaling(GO:0009757) sugar mediated signaling pathway(GO:0010182) glucose mediated signaling pathway(GO:0010255) carbohydrate homeostasis(GO:0033500) glucose homeostasis(GO:0042593) cellular response to monosaccharide stimulus(GO:0071326) cellular response to hexose stimulus(GO:0071331) cellular response to glucose stimulus(GO:0071333) |
| 0.6 | 9.1 | GO:0015833 | peptide transport(GO:0015833) |
| 0.6 | 5.8 | GO:0045053 | protein retention in Golgi apparatus(GO:0045053) |
| 0.6 | 4.7 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.6 | 0.6 | GO:0071902 | positive regulation of protein serine/threonine kinase activity(GO:0071902) |
| 0.6 | 1.1 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.5 | 2.6 | GO:0046656 | folic acid metabolic process(GO:0046655) folic acid biosynthetic process(GO:0046656) |
| 0.5 | 13.0 | GO:0097384 | ergosterol biosynthetic process(GO:0006696) phytosteroid biosynthetic process(GO:0016129) cellular alcohol biosynthetic process(GO:0044108) cellular lipid biosynthetic process(GO:0097384) |
| 0.5 | 3.7 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.5 | 0.9 | GO:0043065 | positive regulation of cell death(GO:0010942) positive regulation of apoptotic process(GO:0043065) positive regulation of programmed cell death(GO:0043068) |
| 0.5 | 1.4 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.4 | 1.8 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.4 | 1.7 | GO:0006596 | polyamine biosynthetic process(GO:0006596) |
| 0.4 | 2.6 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.4 | 2.6 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 0.4 | 0.4 | GO:0061418 | regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061418) |
| 0.4 | 1.7 | GO:0048313 | Golgi inheritance(GO:0048313) |
| 0.4 | 1.6 | GO:0016036 | cellular response to phosphate starvation(GO:0016036) |
| 0.4 | 3.1 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.4 | 2.7 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.4 | 7.5 | GO:0046785 | microtubule nucleation(GO:0007020) microtubule polymerization(GO:0046785) |
| 0.4 | 2.6 | GO:0006639 | neutral lipid metabolic process(GO:0006638) acylglycerol metabolic process(GO:0006639) triglyceride metabolic process(GO:0006641) |
| 0.4 | 2.6 | GO:0010696 | positive regulation of spindle pole body separation(GO:0010696) |
| 0.4 | 3.7 | GO:0001402 | signal transduction involved in filamentous growth(GO:0001402) |
| 0.4 | 3.7 | GO:0034221 | cell wall chitin biosynthetic process(GO:0006038) fungal-type cell wall chitin biosynthetic process(GO:0034221) fungal-type cell wall polysaccharide biosynthetic process(GO:0051278) fungal-type cell wall polysaccharide metabolic process(GO:0071966) |
| 0.4 | 2.2 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.4 | 3.3 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.4 | 0.7 | GO:0033262 | regulation of nuclear cell cycle DNA replication(GO:0033262) |
| 0.4 | 1.8 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.3 | 3.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 2.4 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.3 | 2.3 | GO:0009312 | trehalose biosynthetic process(GO:0005992) oligosaccharide biosynthetic process(GO:0009312) disaccharide biosynthetic process(GO:0046351) |
| 0.3 | 2.9 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.3 | 0.3 | GO:0043966 | histone H3 acetylation(GO:0043966) histone H3-K9 acetylation(GO:0043970) histone H3-K14 acetylation(GO:0044154) histone H3-K9 modification(GO:0061647) |
| 0.3 | 3.2 | GO:0007109 | obsolete cytokinesis, completion of separation(GO:0007109) |
| 0.3 | 21.7 | GO:0006913 | nucleocytoplasmic transport(GO:0006913) |
| 0.3 | 1.9 | GO:0006771 | riboflavin metabolic process(GO:0006771) riboflavin biosynthetic process(GO:0009231) |
| 0.3 | 1.5 | GO:0006658 | phosphatidylserine metabolic process(GO:0006658) |
| 0.3 | 2.9 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.3 | 0.3 | GO:0006952 | defense response(GO:0006952) |
| 0.3 | 4.2 | GO:0000011 | vacuole inheritance(GO:0000011) |
| 0.3 | 0.8 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.3 | 3.1 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.2 | 6.7 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.2 | 4.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 2.1 | GO:0055074 | cellular calcium ion homeostasis(GO:0006874) calcium ion homeostasis(GO:0055074) |
| 0.2 | 0.7 | GO:0035459 | cargo loading into vesicle(GO:0035459) cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.2 | 2.1 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.2 | 0.7 | GO:0000433 | negative regulation of transcription from RNA polymerase II promoter by glucose(GO:0000433) negative regulation of transcription by glucose(GO:0045014) |
| 0.2 | 0.7 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.2 | 0.9 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.2 | 1.1 | GO:0006816 | calcium ion transport(GO:0006816) |
| 0.2 | 4.1 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.2 | 0.7 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.2 | 1.6 | GO:0006672 | ceramide metabolic process(GO:0006672) ceramide biosynthetic process(GO:0046513) |
| 0.2 | 2.0 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.2 | 3.1 | GO:0007094 | mitotic spindle assembly checkpoint(GO:0007094) spindle assembly checkpoint(GO:0071173) |
| 0.2 | 0.7 | GO:0038032 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.2 | 1.7 | GO:0006797 | polyphosphate metabolic process(GO:0006797) |
| 0.2 | 0.4 | GO:0048211 | Golgi vesicle docking(GO:0048211) |
| 0.2 | 2.1 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.2 | 6.2 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.2 | 4.0 | GO:0006414 | translational elongation(GO:0006414) |
| 0.2 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 0.6 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.2 | 1.7 | GO:0006547 | histidine biosynthetic process(GO:0000105) histidine metabolic process(GO:0006547) imidazole-containing compound metabolic process(GO:0052803) |
| 0.2 | 3.4 | GO:0016575 | histone deacetylation(GO:0016575) |
| 0.2 | 2.1 | GO:0051668 | localization within membrane(GO:0051668) |
| 0.2 | 0.7 | GO:0015809 | arginine transport(GO:0015809) |
| 0.2 | 0.3 | GO:0033239 | negative regulation of cellular amine metabolic process(GO:0033239) |
| 0.2 | 2.8 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.2 | 3.0 | GO:0006915 | apoptotic process(GO:0006915) cell death(GO:0008219) programmed cell death(GO:0012501) |
| 0.2 | 1.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.2 | 1.7 | GO:0007009 | plasma membrane organization(GO:0007009) |
| 0.2 | 0.5 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.2 | 0.6 | GO:1901983 | regulation of histone acetylation(GO:0035065) positive regulation of histone acetylation(GO:0035066) regulation of protein acetylation(GO:1901983) positive regulation of protein acetylation(GO:1901985) regulation of peptidyl-lysine acetylation(GO:2000756) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.1 | 1.0 | GO:0016126 | steroid biosynthetic process(GO:0006694) sterol biosynthetic process(GO:0016126) |
| 0.1 | 3.1 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.1 | 1.0 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.1 | 0.4 | GO:0072353 | cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.1 | 1.6 | GO:0007120 | axial cellular bud site selection(GO:0007120) |
| 0.1 | 0.9 | GO:0031578 | mitotic spindle orientation checkpoint(GO:0031578) |
| 0.1 | 0.7 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.1 | 1.6 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.1 | 0.9 | GO:0006544 | glycine metabolic process(GO:0006544) |
| 0.1 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.1 | 0.6 | GO:0006057 | cell wall mannoprotein biosynthetic process(GO:0000032) mannoprotein metabolic process(GO:0006056) mannoprotein biosynthetic process(GO:0006057) cell wall glycoprotein biosynthetic process(GO:0031506) |
| 0.1 | 0.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.1 | 0.7 | GO:0032042 | mitochondrial DNA metabolic process(GO:0032042) |
| 0.1 | 2.1 | GO:0006885 | regulation of pH(GO:0006885) |
| 0.1 | 1.8 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.1 | 0.5 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 1.4 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) pteridine-containing compound biosynthetic process(GO:0042559) |
| 0.1 | 0.5 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.5 | GO:0000755 | cytogamy(GO:0000755) |
| 0.1 | 0.8 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.1 | 0.3 | GO:0015940 | pantothenate metabolic process(GO:0015939) pantothenate biosynthetic process(GO:0015940) |
| 0.1 | 0.2 | GO:0006026 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.1 | 0.3 | GO:0055129 | proline metabolic process(GO:0006560) proline biosynthetic process(GO:0006561) L-proline biosynthetic process(GO:0055129) |
| 0.1 | 0.2 | GO:1901977 | regulation of cell cycle checkpoint(GO:1901976) negative regulation of cell cycle checkpoint(GO:1901977) regulation of DNA damage checkpoint(GO:2000001) negative regulation of DNA damage checkpoint(GO:2000002) negative regulation of response to DNA damage stimulus(GO:2001021) |
| 0.1 | 0.2 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.1 | 0.2 | GO:0006037 | cell wall chitin metabolic process(GO:0006037) |
| 0.1 | 0.6 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 0.4 | GO:0000279 | mitotic M phase(GO:0000087) M phase(GO:0000279) |
| 0.1 | 2.2 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.1 | 0.2 | GO:0010907 | positive regulation of glucose metabolic process(GO:0010907) positive regulation of gluconeogenesis(GO:0045722) |
| 0.1 | 0.5 | GO:0016233 | telomere capping(GO:0016233) |
| 0.1 | 0.3 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.1 | 0.1 | GO:2000222 | positive regulation of cell growth(GO:0030307) positive regulation of pseudohyphal growth by positive regulation of transcription from RNA polymerase II promoter(GO:1900461) positive regulation of pseudohyphal growth(GO:2000222) |
| 0.0 | 0.2 | GO:0032272 | negative regulation of actin filament polymerization(GO:0030837) negative regulation of protein polymerization(GO:0032272) |
| 0.0 | 0.6 | GO:0010970 | establishment of localization by movement along microtubule(GO:0010970) nuclear migration along microtubule(GO:0030473) organelle transport along microtubule(GO:0072384) |
| 0.0 | 0.3 | GO:0016125 | steroid metabolic process(GO:0008202) sterol metabolic process(GO:0016125) |
| 0.0 | 0.4 | GO:0006614 | cotranslational protein targeting to membrane(GO:0006613) SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.5 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0019217 | regulation of fatty acid metabolic process(GO:0019217) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) positive regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0090282) |
| 0.0 | 1.0 | GO:0051123 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.1 | GO:0061408 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) positive regulation of transcription from RNA polymerase II promoter in response to heat stress(GO:0061408) |
| 0.0 | 0.6 | GO:0006896 | Golgi to vacuole transport(GO:0006896) |
| 0.0 | 0.4 | GO:0045454 | cell redox homeostasis(GO:0045454) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.6 | 25.9 | GO:0070545 | PeBoW complex(GO:0070545) |
| 7.3 | 36.3 | GO:0030689 | Noc complex(GO:0030689) |
| 6.2 | 24.7 | GO:0031429 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 6.1 | 24.5 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 6.0 | 84.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 5.1 | 30.7 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 5.0 | 221.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 5.0 | 19.9 | GO:0070860 | RNA polymerase I core factor complex(GO:0070860) |
| 4.8 | 14.5 | GO:0034457 | Mpp10 complex(GO:0034457) |
| 4.8 | 38.4 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 4.8 | 14.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 4.7 | 28.2 | GO:0034455 | t-UTP complex(GO:0034455) |
| 3.6 | 80.2 | GO:0032040 | small-subunit processome(GO:0032040) |
| 3.4 | 16.9 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 3.3 | 9.8 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 3.2 | 19.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 3.1 | 9.4 | GO:0097344 | Rix1 complex(GO:0097344) |
| 2.9 | 11.6 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 2.4 | 24.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 2.4 | 7.1 | GO:0005854 | nascent polypeptide-associated complex(GO:0005854) |
| 2.3 | 88.3 | GO:0030684 | preribosome(GO:0030684) |
| 2.2 | 8.9 | GO:0034448 | EGO complex(GO:0034448) |
| 2.2 | 6.6 | GO:0008275 | gamma-tubulin small complex, spindle pole body(GO:0000928) gamma-tubulin complex(GO:0000930) gamma-tubulin small complex(GO:0008275) |
| 2.1 | 279.1 | GO:0005730 | nucleolus(GO:0005730) |
| 1.9 | 7.7 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 1.8 | 11.0 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 1.7 | 8.7 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 1.7 | 6.9 | GO:0005946 | alpha,alpha-trehalose-phosphate synthase complex (UDP-forming)(GO:0005946) |
| 1.6 | 4.9 | GO:0043529 | GET complex(GO:0043529) |
| 1.6 | 4.7 | GO:0031417 | NatC complex(GO:0031417) |
| 1.6 | 6.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 1.5 | 9.3 | GO:0032126 | eisosome(GO:0032126) |
| 1.5 | 5.9 | GO:0033597 | mitotic checkpoint complex(GO:0033597) |
| 1.4 | 7.0 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 1.4 | 4.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 1.3 | 98.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 1.2 | 3.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 1.1 | 5.6 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 1.1 | 5.4 | GO:0030478 | actin cap(GO:0030478) |
| 1.1 | 3.2 | GO:0035649 | Nrd1 complex(GO:0035649) |
| 0.9 | 2.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.9 | 32.4 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.9 | 2.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.9 | 2.6 | GO:0051286 | cell tip(GO:0051286) |
| 0.8 | 4.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.8 | 5.0 | GO:0005880 | nuclear microtubule(GO:0005880) |
| 0.8 | 2.4 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.8 | 2.3 | GO:0017102 | methionyl glutamyl tRNA synthetase complex(GO:0017102) |
| 0.7 | 2.9 | GO:0000444 | MIS12/MIND type complex(GO:0000444) nuclear MIS12/MIND complex(GO:0000818) |
| 0.7 | 2.2 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.7 | 9.8 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.7 | 2.1 | GO:0005845 | mRNA cap binding complex(GO:0005845) |
| 0.7 | 0.7 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.6 | 6.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.6 | 4.3 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.6 | 13.5 | GO:0005844 | polysome(GO:0005844) |
| 0.6 | 4.7 | GO:0042597 | periplasmic space(GO:0042597) |
| 0.6 | 4.0 | GO:0033180 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.6 | 1.1 | GO:0030428 | cell septum(GO:0030428) |
| 0.5 | 2.0 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.5 | 1.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.5 | 2.9 | GO:0070772 | PAS complex(GO:0070772) |
| 0.5 | 1.8 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.4 | 6.3 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.4 | 2.7 | GO:0035339 | serine C-palmitoyltransferase complex(GO:0017059) SPOTS complex(GO:0035339) |
| 0.4 | 2.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.4 | 1.8 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.4 | 1.7 | GO:0033254 | vacuolar transporter chaperone complex(GO:0033254) |
| 0.4 | 9.5 | GO:0030173 | integral component of Golgi membrane(GO:0030173) intrinsic component of Golgi membrane(GO:0031228) |
| 0.4 | 2.8 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.4 | 3.0 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 4.5 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.4 | 1.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.3 | 4.3 | GO:0005940 | septin ring(GO:0005940) |
| 0.3 | 1.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.3 | 11.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.3 | 0.3 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.3 | 7.7 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.2 | 0.2 | GO:0071819 | DUBm complex(GO:0071819) |
| 0.2 | 0.7 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.2 | 6.1 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.2 | 20.7 | GO:0005759 | mitochondrial matrix(GO:0005759) |
| 0.2 | 4.3 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.2 | 2.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.2 | 2.5 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.2 | 0.8 | GO:0097042 | extrinsic component of fungal-type vacuolar membrane(GO:0097042) |
| 0.2 | 3.4 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 2.0 | GO:0000243 | commitment complex(GO:0000243) |
| 0.2 | 0.6 | GO:0044697 | HICS complex(GO:0044697) |
| 0.2 | 2.7 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.2 | 3.7 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.2 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.2 | 0.9 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 1.0 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.2 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.2 | 0.6 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.1 | 0.6 | GO:0000817 | COMA complex(GO:0000817) |
| 0.1 | 0.9 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.4 | GO:0032301 | MutSalpha complex(GO:0032301) |
| 0.1 | 1.0 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.1 | 2.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.1 | 15.0 | GO:0005840 | ribosome(GO:0005840) |
| 0.1 | 4.9 | GO:0044445 | cytosolic part(GO:0044445) |
| 0.1 | 0.7 | GO:0033101 | cellular bud membrane(GO:0033101) |
| 0.1 | 1.6 | GO:0016586 | RSC complex(GO:0016586) |
| 0.1 | 0.8 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) BAF-type complex(GO:0090544) |
| 0.1 | 0.3 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.1 | 0.6 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 0.2 | GO:0070823 | HDA1 complex(GO:0070823) |
| 0.1 | 1.7 | GO:0005887 | integral component of plasma membrane(GO:0005887) |
| 0.1 | 2.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.6 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.1 | 0.6 | GO:0070938 | cellular bud neck contractile ring(GO:0000142) actomyosin contractile ring(GO:0005826) contractile ring(GO:0070938) |
| 0.1 | 0.4 | GO:0030867 | rough endoplasmic reticulum(GO:0005791) rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.4 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.4 | GO:0046695 | SLIK (SAGA-like) complex(GO:0046695) |
| 0.0 | 0.6 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000417 | HIR complex(GO:0000417) |
| 0.0 | 1.0 | GO:0000131 | incipient cellular bud site(GO:0000131) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0009898 | cytoplasmic side of plasma membrane(GO:0009898) |
| 0.0 | 0.1 | GO:0036452 | ESCRT complex(GO:0036452) |
| 0.0 | 0.0 | GO:0097196 | Shu complex(GO:0097196) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.2 | 18.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 6.2 | 6.2 | GO:0033592 | RNA strand annealing activity(GO:0033592) annealing activity(GO:0097617) |
| 6.1 | 18.2 | GO:0001169 | transcription factor activity, RNA polymerase I CORE element sequence-specific binding(GO:0001169) transcription factor activity, RNA polymerase I CORE element binding transcription factor recruiting(GO:0001187) |
| 6.0 | 84.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 5.7 | 17.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 5.6 | 111.7 | GO:0030515 | snoRNA binding(GO:0030515) |
| 5.6 | 16.7 | GO:0019003 | GDP binding(GO:0019003) |
| 4.5 | 13.5 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 4.4 | 13.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 4.2 | 12.5 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 4.0 | 8.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 3.7 | 11.2 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 3.6 | 14.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 3.4 | 17.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 3.2 | 19.4 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 3.1 | 12.5 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 2.9 | 2.9 | GO:0001082 | transcription factor activity, RNA polymerase I transcription factor binding(GO:0001082) |
| 2.9 | 5.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 2.9 | 8.7 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 2.9 | 88.6 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 2.7 | 8.2 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 2.7 | 10.8 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 2.6 | 7.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 2.6 | 15.8 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 2.4 | 7.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 2.4 | 24.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 2.4 | 14.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 2.3 | 6.9 | GO:0003825 | alpha,alpha-trehalose-phosphate synthase (UDP-forming) activity(GO:0003825) |
| 2.2 | 6.6 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 2.1 | 6.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 2.1 | 6.2 | GO:0047066 | phospholipid-hydroperoxide glutathione peroxidase activity(GO:0047066) |
| 2.0 | 6.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 2.0 | 8.1 | GO:0000297 | spermine transmembrane transporter activity(GO:0000297) |
| 2.0 | 14.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 1.9 | 7.7 | GO:0016725 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 1.9 | 9.6 | GO:0008186 | RNA-dependent ATPase activity(GO:0008186) |
| 1.8 | 5.4 | GO:0004326 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) |
| 1.7 | 15.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 1.7 | 17.0 | GO:0019239 | deaminase activity(GO:0019239) |
| 1.7 | 5.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 1.6 | 1.6 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 1.6 | 4.9 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 1.6 | 4.9 | GO:0015166 | polyol transmembrane transporter activity(GO:0015166) |
| 1.5 | 4.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 1.5 | 6.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
| 1.5 | 13.1 | GO:0004497 | monooxygenase activity(GO:0004497) |
| 1.4 | 12.5 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 1.4 | 8.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 1.4 | 23.4 | GO:0016763 | transferase activity, transferring pentosyl groups(GO:0016763) |
| 1.4 | 52.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 1.4 | 5.4 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 1.3 | 18.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 1.3 | 6.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 1.3 | 3.8 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 1.2 | 5.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 1.2 | 3.7 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 1.2 | 3.6 | GO:0004100 | chitin synthase activity(GO:0004100) |
| 1.2 | 5.9 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 1.2 | 4.7 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 1.2 | 9.3 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 1.2 | 5.8 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 1.1 | 10.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 1.0 | 5.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 1.0 | 4.9 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 1.0 | 5.8 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 1.0 | 4.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 1.0 | 2.9 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.9 | 2.8 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.9 | 75.2 | GO:0032561 | GTP binding(GO:0005525) guanyl nucleotide binding(GO:0019001) guanyl ribonucleotide binding(GO:0032561) |
| 0.9 | 2.7 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.9 | 3.5 | GO:0004338 | glucan exo-1,3-beta-glucosidase activity(GO:0004338) |
| 0.9 | 3.4 | GO:0008173 | RNA methyltransferase activity(GO:0008173) |
| 0.9 | 2.6 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.8 | 4.2 | GO:0016408 | C-acyltransferase activity(GO:0016408) |
| 0.8 | 3.3 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.8 | 140.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.8 | 7.8 | GO:0019205 | nucleobase-containing compound kinase activity(GO:0019205) |
| 0.8 | 9.9 | GO:0000049 | tRNA binding(GO:0000049) |
| 0.8 | 187.7 | GO:0003723 | RNA binding(GO:0003723) |
| 0.7 | 6.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.7 | 4.4 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.7 | 26.2 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.7 | 12.7 | GO:0042803 | protein homodimerization activity(GO:0042803) |
| 0.7 | 3.4 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.7 | 2.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.6 | 5.2 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.6 | 3.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.6 | 2.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.6 | 0.6 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.6 | 9.4 | GO:0005506 | iron ion binding(GO:0005506) |
| 0.6 | 2.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.6 | 5.9 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.6 | 1.8 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.6 | 5.5 | GO:0001133 | RNA polymerase II transcription factor activity, sequence-specific transcription regulatory region DNA binding(GO:0001133) |
| 0.5 | 5.3 | GO:0001083 | transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) |
| 0.5 | 2.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.5 | 4.1 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.5 | 3.0 | GO:0004529 | exodeoxyribonuclease activity(GO:0004529) |
| 0.5 | 2.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.5 | 1.5 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
| 0.5 | 2.0 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.5 | 6.6 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.5 | 5.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.5 | 3.7 | GO:0005310 | dicarboxylic acid transmembrane transporter activity(GO:0005310) |
| 0.5 | 0.9 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.4 | 4.9 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.4 | 4.4 | GO:0005496 | steroid binding(GO:0005496) sterol binding(GO:0032934) |
| 0.4 | 12.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.4 | 4.1 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.4 | 1.6 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.4 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.3 | 3.0 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.3 | 1.0 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.3 | 0.6 | GO:0016840 | carbon-nitrogen lyase activity(GO:0016840) |
| 0.3 | 1.3 | GO:0008252 | nucleotidase activity(GO:0008252) |
| 0.3 | 2.5 | GO:0010181 | FMN binding(GO:0010181) |
| 0.3 | 4.6 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.3 | 1.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.3 | 5.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.3 | 1.7 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.3 | 2.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.3 | 1.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.3 | 1.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.3 | 2.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.3 | 1.1 | GO:0016655 | oxidoreductase activity, acting on NAD(P)H, quinone or similar compound as acceptor(GO:0016655) |
| 0.3 | 1.3 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.3 | 2.9 | GO:0034061 | DNA-directed DNA polymerase activity(GO:0003887) DNA polymerase activity(GO:0034061) |
| 0.3 | 1.8 | GO:0051920 | thioredoxin peroxidase activity(GO:0008379) peroxiredoxin activity(GO:0051920) |
| 0.2 | 5.8 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.2 | 3.4 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.2 | 1.2 | GO:0034318 | alcohol O-acetyltransferase activity(GO:0004026) alcohol O-acyltransferase activity(GO:0034318) |
| 0.2 | 0.7 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.2 | 0.7 | GO:0008144 | drug binding(GO:0008144) |
| 0.2 | 1.8 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.2 | 0.9 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.2 | 0.8 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.2 | 4.5 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 1.0 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) phosphatidylinositol transporter activity(GO:0008526) |
| 0.2 | 1.2 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.2 | 1.7 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.2 | 0.5 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.2 | 0.2 | GO:0004488 | methylenetetrahydrofolate dehydrogenase (NADP+) activity(GO:0004488) |
| 0.2 | 11.0 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.2 | 1.0 | GO:0008408 | 3'-5' exonuclease activity(GO:0008408) |
| 0.1 | 4.6 | GO:0046983 | protein dimerization activity(GO:0046983) |
| 0.1 | 1.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.1 | 4.9 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.1 | 3.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 10.0 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.1 | 1.2 | GO:0000009 | alpha-1,6-mannosyltransferase activity(GO:0000009) |
| 0.1 | 1.3 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.1 | 0.5 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.1 | 2.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.1 | 0.6 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.1 | 1.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.5 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 0.3 | GO:0016642 | glycine dehydrogenase (decarboxylating) activity(GO:0004375) oxidoreductase activity, acting on the CH-NH2 group of donors, disulfide as acceptor(GO:0016642) |
| 0.1 | 1.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 2.0 | GO:0016667 | oxidoreductase activity, acting on a sulfur group of donors(GO:0016667) |
| 0.1 | 2.9 | GO:0005085 | guanyl-nucleotide exchange factor activity(GO:0005085) |
| 0.1 | 0.4 | GO:0070546 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.1 | 1.3 | GO:0005048 | signal sequence binding(GO:0005048) |
| 0.1 | 0.2 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.1 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.7 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 1.4 | GO:0008757 | S-adenosylmethionine-dependent methyltransferase activity(GO:0008757) |
| 0.1 | 0.2 | GO:0001128 | RNA polymerase II transcription coactivator activity involved in preinitiation complex assembly(GO:0001128) |
| 0.1 | 0.2 | GO:0005034 | osmosensor activity(GO:0005034) |
| 0.1 | 0.8 | GO:0015926 | glucosidase activity(GO:0015926) |
| 0.1 | 0.4 | GO:1901618 | organic hydroxy compound transmembrane transporter activity(GO:1901618) |
| 0.1 | 0.4 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 0.1 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0015088 | copper uptake transmembrane transporter activity(GO:0015088) |
| 0.0 | 0.2 | GO:0042123 | glucanosyltransferase activity(GO:0042123) 1,3-beta-glucanosyltransferase activity(GO:0042124) |
| 0.0 | 0.1 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 0.1 | GO:0016651 | oxidoreductase activity, acting on NAD(P)H(GO:0016651) |
| 0.0 | 0.3 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.6 | GO:0008565 | protein transporter activity(GO:0008565) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.3 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 1.5 | 4.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 1.5 | 1.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 1.5 | 5.9 | PID P73PATHWAY | p73 transcription factor network |
| 1.4 | 5.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.6 | 2.5 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.4 | 1.3 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.3 | 0.7 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.3 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 0.4 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.1 | 0.4 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.1 | 0.4 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.8 | 11.1 | REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS | Genes involved in Glycerophospholipid biosynthesis |
| 2.0 | 16.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 2.0 | 14.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 1.8 | 14.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 1.7 | 5.0 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 1.2 | 19.7 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 1.1 | 2.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 1.0 | 5.9 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.7 | 5.8 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.6 | 0.6 | REACTOME METABOLISM OF MRNA | Genes involved in Metabolism of mRNA |
| 0.6 | 1.2 | REACTOME ACTIVATED TLR4 SIGNALLING | Genes involved in Activated TLR4 signalling |
| 0.5 | 2.5 | REACTOME APOPTOSIS | Genes involved in Apoptosis |
| 0.4 | 0.4 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.3 | 2.0 | REACTOME PROTEIN FOLDING | Genes involved in Protein folding |
| 0.3 | 3.6 | REACTOME TRANSCRIPTION | Genes involved in Transcription |
| 0.2 | 1.5 | REACTOME AXON GUIDANCE | Genes involved in Axon guidance |
| 0.1 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 0.7 | REACTOME SIGNALING BY GPCR | Genes involved in Signaling by GPCR |
| 0.1 | 0.2 | REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | Genes involved in Fatty acid, triacylglycerol, and ketone body metabolism |
| 0.0 | 0.2 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |