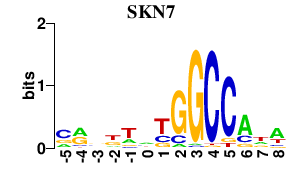
Results for SKN7
Z-value: 0.38
Transcription factors associated with SKN7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
SKN7
|
S000001249 | Nuclear response regulator and transcription factor |
Activity-expression correlation:
Activity profile of SKN7 motif
Sorted Z-values of SKN7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| YGL177W | 0.90 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YAR053W | 0.84 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YPL230W | 0.49 |
USV1
|
Putative transcription factor containing a C2H2 zinc finger; mutation affects transcriptional regulation of genes involved in protein folding, ATP binding, and cell wall biosynthesis |
|
| YGR087C | 0.47 |
PDC6
|
Minor isoform of pyruvate decarboxylase, decarboxylates pyruvate to acetaldehyde, involved in amino acid catabolism; transcription is glucose- and ethanol-dependent, and is strongly induced during sulfur limitation |
|
| YBR157C | 0.47 |
ICS2
|
Protein of unknown function; null mutation does not confer any obvious defects in growth, spore germination, viability, or carbohydrate utilization |
|
| YOR186W | 0.45 |
Putative protein of unknown function; proper regulation of expression during heat stress is sphingolipid-dependent |
||
| YBR296C | 0.44 |
PHO89
|
Na+/Pi cotransporter, active in early growth phase; similar to phosphate transporters of Neurospora crassa; transcription regulated by inorganic phosphate concentrations and Pho4p |
|
| YBR292C | 0.43 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YBR292C is not an essential gene |
||
| YOL052C-A | 0.40 |
DDR2
|
Multistress response protein, expression is activated by a variety of xenobiotic agents and environmental or physiological stresses |
|
| YLR164W | 0.38 |
Mitochondrial inner membrane of unknown function; similar to Tim18p and Sdh4p; expression induced by nitrogen limitation in a GLN3, GAT1-dependent manner |
||
| YLR122C | 0.37 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR123C |
||
| YGR051C | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; YGR051C is not an essential gene |
||
| YJR128W | 0.36 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the verified ORF RSF2 |
||
| YIR038C | 0.34 |
GTT1
|
ER associated glutathione S-transferase capable of homodimerization; expression induced during the diauxic shift and throughout stationary phase; functional overlap with Gtt2p, Grx1p, and Grx2p |
|
| YPL026C | 0.34 |
SKS1
|
Putative serine/threonine protein kinase; involved in the adaptation to low concentrations of glucose independent of the SNF3 regulated pathway |
|
| YLR123C | 0.32 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YLR122C; contains characteristic aminoacyl-tRNA motif |
||
| YGR236C | 0.31 |
SPG1
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YLR124W | 0.31 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YNL179C | 0.31 |
Dubious open reading frame, unlikely to encode a protein; not conserved in closely related Saccharomyces species; deletion in cyr1 mutant results in loss of stress resistance |
||
| YHR033W | 0.31 |
Putative protein of unknown function; epitope-tagged protein localizes to the cytoplasm |
||
| YHR048W | 0.29 |
YHK8
|
Presumed antiporter of the DHA1 family of multidrug resistance transporters; contains 12 predicted transmembrane spans; expression of gene is up-regulated in cells exhibiting reduced susceptibility to azoles |
|
| YNL180C | 0.28 |
RHO5
|
Non-essential small GTPase of the Rho/Rac subfamily of Ras-like proteins, likely involved in protein kinase C (Pkc1p)-dependent signal transduction pathway that controls cell integrity |
|
| YGR039W | 0.27 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the Autonomously Replicating Sequence ARS722 |
||
| YLR162W-A | 0.25 |
Putative protein of unknown function identified by fungal homology comparisons and RT-PCR |
||
| YOR348C | 0.25 |
PUT4
|
Proline permease, required for high-affinity transport of proline; also transports the toxic proline analog azetidine-2-carboxylate (AzC); PUT4 transcription is repressed in ammonia-grown cells |
|
| YKL165C-A | 0.24 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR088W | 0.23 |
CTT1
|
Cytosolic catalase T, has a role in protection from oxidative damage by hydrogen peroxide |
|
| YGL035C | 0.23 |
MIG1
|
Transcription factor involved in glucose repression; sequence specific DNA binding protein containing two Cys2His2 zinc finger motifs; regulated by the SNF1 kinase and the GLC7 phosphatase |
|
| YLR337C | 0.22 |
VRP1
|
Proline-rich actin-associated protein involved in cytoskeletal organization and cytokinesis; related to mammalian Wiskott-Aldrich syndrome protein (WASP)-interacting protein (WIP) |
|
| YNL091W | 0.22 |
NST1
|
Protein of unknown function, mediates sensitivity to salt stress; interacts physically with the splicing factor Msl1p and also displays genetic interaction with MSL1 |
|
| YDR536W | 0.22 |
STL1
|
Glycerol proton symporter of the plasma membrane, subject to glucose-induced inactivation, strongly but transiently induced when cells are subjected to osmotic shock |
|
| YJR120W | 0.20 |
Protein of unknown function; essential for growth under anaerobic conditions; mutation causes decreased expression of ATP2, impaired respiration, defective sterol uptake, and altered levels/localization of ABC transporters Aus1p and Pdr11p |
||
| YLR307W | 0.20 |
CDA1
|
Chitin deacetylase, together with Cda2p involved in the biosynthesis ascospore wall component, chitosan; required for proper rigidity of the ascospore wall |
|
| YNR002C | 0.20 |
ATO2
|
Putative transmembrane protein involved in export of ammonia, a starvation signal that promotes cell death in aging colonies; phosphorylated in mitochondria; member of the TC 9.B.33 YaaH family; homolog of Ady2p and Y. lipolytica Gpr1p |
|
| YAR070C | 0.20 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YLR154W-A | 0.20 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YJR150C | 0.20 |
DAN1
|
Cell wall mannoprotein with similarity to Tir1p, Tir2p, Tir3p, and Tir4p; expressed under anaerobic conditions, completely repressed during aerobic growth |
|
| YPR151C | 0.19 |
SUE1
|
Mitochondrial protein required for degradation of unstable forms of cytochrome c |
|
| YBR291C | 0.19 |
CTP1
|
Mitochondrial inner membrane citrate transporter, member of the mitochondrial carrier family |
|
| YOR072W-A | 0.19 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YOR072W; originally identified by fungal homology and RT-PCR |
||
| YPL024W | 0.19 |
RMI1
|
Involved in response to DNA damage; null mutants have increased rates of recombination and delayed S phase; interacts physically and genetically with Sgs1p (RecQ family member) and Top3p (topoisomerase III) |
|
| YDR119W-A | 0.18 |
Putative protein of unknown function |
||
| YOR032W-A | 0.18 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YIL136W | 0.17 |
OM45
|
Protein of unknown function, major constituent of the mitochondrial outer membrane; located on the outer (cytosolic) face of the outer membrane |
|
| YOR072W | 0.17 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the dubious gene YOR072W-A; diploid deletion strains are methotrexate, paraquat and wortmannin sensitive |
||
| YHR032W-A | 0.17 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YHR032C-A |
||
| YIR019C | 0.17 |
MUC1
|
GPI-anchored cell surface glycoprotein (flocculin) required for pseudohyphal formation, invasive growth, flocculation, and biofilms; transcriptionally regulated by the MAPK pathway (via Ste12p and Tec1p) and the cAMP pathway (via Flo8p) |
|
| YGR259C | 0.16 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps almost completely with the verified ORF TNA1/YGR260W |
||
| YGR258C | 0.16 |
RAD2
|
Single-stranded DNA endonuclease, cleaves single-stranded DNA during nucleotide excision repair to excise damaged DNA; subunit of Nucleotide Excision Repair Factor 3 (NEF3); homolog of human XPG protein |
|
| YAL062W | 0.16 |
GDH3
|
NADP(+)-dependent glutamate dehydrogenase, synthesizes glutamate from ammonia and alpha-ketoglutarate; rate of alpha-ketoglutarate utilization differs from Gdh1p; expression regulated by nitrogen and carbon sources |
|
| YGL033W | 0.16 |
HOP2
|
Meiosis-specific protein that localizes to chromosomes, preventing synapsis between nonhomologous chromosomes and ensuring synapsis between homologs; complexes with Mnd1p to promote homolog pairing and meiotic double-strand break repair |
|
| YIL099W | 0.15 |
SGA1
|
Intracellular sporulation-specific glucoamylase involved in glycogen degradation; induced during starvation of a/a diploids late in sporulation, but dispensable for sporulation |
|
| YMR107W | 0.14 |
SPG4
|
Protein required for survival at high temperature during stationary phase; not required for growth on nonfermentable carbon sources |
|
| YOR186C-A | 0.14 |
Identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YLL060C | 0.14 |
GTT2
|
Glutathione S-transferase capable of homodimerization; functional overlap with Gtt2p, Grx1p, and Grx2p |
|
| YGR260W | 0.14 |
TNA1
|
High affinity nicotinic acid plasma membrane permease, responsible for uptake of low levels of nicotinic acid; expression of the gene increases in the absence of extracellular nicotinic acid or para-aminobenzoate (PABA) |
|
| YOR378W | 0.14 |
Putative protein of unknown function; belongs to the DHA2 family of drug:H+ antiporters; YOR378W is not an essential gene |
||
| YPL261C | 0.14 |
Dubious open reading frame unlikely to encode a protein; partially overlaps the uncharacterized ORF YPL260W |
||
| YPR077C | 0.13 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression increased by deletion of NAP1 |
||
| YMR114C | 0.13 |
Protein of unknown function; may interact with ribosomes, based on co-purification experiments; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and cytoplasm; YMR114C is not an essential gene |
||
| YPR030W | 0.13 |
CSR2
|
Nuclear protein with a potential regulatory role in utilization of galactose and nonfermentable carbon sources; overproduction suppresses the lethality at high temperature of a chs5 spa2 double null mutation; potential Cdc28p substrate |
|
| YMR165C | 0.13 |
PAH1
|
Mg2+-dependent phosphatidate (PA) phosphatase, catalyzes the dephosphorylation of PA to yield diacylglycerol and Pi, responsible for de novo lipid synthesis; homologous to mammalian lipin 1 |
|
| YEL049W | 0.13 |
PAU2
|
Part of 23-member seripauperin multigene family encoded mainly in subtelomeric regions, active during alcoholic fermentation, regulated by anaerobiosis, negatively regulated by oxygen, repressed by heme |
|
| YKR011C | 0.13 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the nucleus |
||
| YIL101C | 0.13 |
XBP1
|
Transcriptional repressor that binds to promoter sequences of the cyclin genes, CYS3, and SMF2; expression is induced by stress or starvation during mitosis, and late in meiosis; member of the Swi4p/Mbp1p family; potential Cdc28p substrate |
|
| YIL045W | 0.13 |
PIG2
|
Putative type-1 protein phosphatase targeting subunit that tethers Glc7p type-1 protein phosphatase to Gsy2p glycogen synthase |
|
| YOR344C | 0.13 |
TYE7
|
Serine-rich protein that contains a basic-helix-loop-helix (bHLH) DNA binding motif; binds E-boxes of glycolytic genes and contributes to their activation; may function as a transcriptional activator in Ty1-mediated gene expression |
|
| YER062C | 0.13 |
HOR2
|
One of two redundant DL-glycerol-3-phosphatases (RHR2/GPP1 encodes the other) involved in glycerol biosynthesis; induced in response to hyperosmotic stress and oxidative stress, and during the diauxic transition |
|
| YBR201C-A | 0.12 |
Putative protein of unknown function |
||
| YOR345C | 0.12 |
Dubious ORF unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the verified gene REV1; null mutant displays increased resistance to antifungal agents gliotoxin, cycloheximide and H2O2 |
||
| YER116C | 0.12 |
SLX8
|
Subunit of the Slx5-Slx8 substrate-specific ubiquitin ligase complex; stimulated by prior attachment of SUMO to the substrate |
|
| YOR066W | 0.12 |
MSA1
|
Activator of G1-specific transcription factors, MBF and SBF, that regulates both the timing of G1-specific gene transcription, and cell cycle initiation; potential Cdc28p substrate |
|
| YDR014W-A | 0.12 |
HED1
|
Meiosis-specific protein that down-regulates Rad51p-mediated mitotic recombination when the meiotic recombination machinery is impaired; early meiotic gene, transcribed specifically during meiotic prophase |
|
| YJL163C | 0.12 |
Putative protein of unknown function |
||
| YBR105C | 0.12 |
VID24
|
Peripheral membrane protein located at Vid (vacuole import and degradation) vesicles; regulates fructose-1,6-bisphosphatase (FBPase) targeting to the vacuole; involved in proteasome-dependent catabolite degradation of FBPase |
|
| YOR071C | 0.12 |
NRT1
|
High-affinity nicotinamide riboside transporter; also transports thiamine with low affinity; shares sequence similarity with Thi7p and Thi72p; proposed to be involved in 5-fluorocytosine sensitivity |
|
| YIL100W | 0.12 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; completely overlaps the dubious ORF YIL100C-A |
||
| YPL061W | 0.12 |
ALD6
|
Cytosolic aldehyde dehydrogenase, activated by Mg2+ and utilizes NADP+ as the preferred coenzyme; required for conversion of acetaldehyde to acetate; constitutively expressed; locates to the mitochondrial outer surface upon oxidative stress |
|
| YDR259C | 0.12 |
YAP6
|
Putative basic leucine zipper (bZIP) transcription factor; overexpression increases sodium and lithium tolerance |
|
| YML089C | 0.12 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data; expression induced by calcium shortage |
||
| YOR185C | 0.11 |
GSP2
|
GTP binding protein (mammalian Ranp homolog) involved in the maintenance of nuclear organization, RNA processing and transport; interacts with Kap121p, Kap123p and Pdr6p (karyophilin betas); Gsp1p homolog that is not required for viability |
|
| YOR032C | 0.11 |
HMS1
|
Basic helix-loop-helix (bHLH) protein with similarity to myc-family transcription factors; overexpression confers hyperfilamentous growth and suppresses the pseudohyphal filamentation defect of a diploid mep1 mep2 homozygous null mutant |
|
| YHR212W-A | 0.11 |
Putative protein of unknown function; identified by gene-trapping, microarray-based expression analysis, and genome-wide homology searching |
||
| YHR211W | 0.11 |
FLO5
|
Lectin-like cell wall protein (flocculin) involved in flocculation, binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin resistant but heat labile; similar to Flo1p |
|
| YER020W | 0.11 |
GPA2
|
Nucleotide binding alpha subunit of the heterotrimeric G protein that interacts with the receptor Gpr1p, has signaling role in response to nutrients; green fluorescent protein (GFP)-fusion protein localizes to the cell periphery |
|
| YLL019C | 0.11 |
KNS1
|
Nonessential putative protein kinase of unknown cellular role; member of the LAMMER family of protein kinases, which are serine/threonine kinases also capable of phosphorylating tyrosine residues |
|
| YDL139C | 0.11 |
SCM3
|
Nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3-H4 histones, required for kinetochore assembly; contains nuclear export signal (NES); required for G2/M progression and localization of Cse4p |
|
| YBR280C | 0.11 |
SAF1
|
F-Box protein involved in proteasome-dependent degradation of Aah1p during entry of cells into quiescence; interacts with Skp1 |
|
| YJR078W | 0.11 |
BNA2
|
Putative tryptophan 2,3-dioxygenase or indoleamine 2,3-dioxygenase, required for the de novo biosynthesis of NAD from tryptophan via kynurenine; expression regulated by Hst1p and Aft2p |
|
| YLR154W-B | 0.11 |
Dubious open reading frame unlikely to encode a protein; encoded within the the 25S rRNA gene on the opposite strand |
||
| YGL163C | 0.11 |
RAD54
|
DNA-dependent ATPase, stimulates strand exchange by modifying the topology of double-stranded DNA; involved in the recombinational repair of double-strand breaks in DNA during vegetative growth and meiosis; member of the SWI/SNF family |
|
| YAR060C | 0.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YBR083W | 0.10 |
TEC1
|
Transcription factor required for full Ty1 epxression, Ty1-mediated gene activation, and haploid invasive and diploid pseudohyphal growth; TEA/ATTS DNA-binding domain family member |
|
| YKL163W | 0.10 |
PIR3
|
O-glycosylated covalently-bound cell wall protein required for cell wall stability; expression is cell cycle regulated, peaking in M/G1 and also subject to regulation by the cell integrity pathway |
|
| YBR222C | 0.10 |
PCS60
|
Peroxisomal AMP-binding protein, localizes to both the peroxisomal peripheral membrane and matrix, expression is highly inducible by oleic acid, similar to E. coli long chain acyl-CoA synthetase |
|
| YBR056W-A | 0.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YBR056C-B |
||
| YDR466W | 0.10 |
PKH3
|
Protein kinase with similarity to mammalian phosphoinositide-dependent kinase 1 (PDK1) and yeast Pkh1p and Pkh2p, two redundant upstream activators of Pkc1p; identified as a multicopy suppressor of a pkh1 pkh2 double mutant |
|
| YDL114W | 0.10 |
Putative protein of unknown function with similarity to acyl-carrier-protein reductases; YDL114W is not an essential gene |
||
| YMR213W | 0.10 |
CEF1
|
Essential splicing factor; associated with Prp19p and the spliceosome, contains an N-terminal c-Myb DNA binding motif necessary for cell viability but not for Prp19p association, evolutionarily conserved and homologous to S. pombe Cdc5p |
|
| YMR206W | 0.10 |
Putative protein of unknown function; YMR206W is not an essential gene |
||
| YIL164C | 0.10 |
NIT1
|
Nitrilase, member of the nitrilase branch of the nitrilase superfamily; in closely related species and other S. cerevisiae strain backgrounds YIL164C and adjacent ORF, YIL165C, likely constitute a single ORF encoding a nitrilase gene |
|
| YJR094C | 0.10 |
IME1
|
Master regulator of meiosis that is active only during meiotic events, activates transcription of early meiotic genes through interaction with Ume6p, degraded by the 26S proteasome following phosphorylation by Ime2p |
|
| YFR016C | 0.10 |
Putative protein of unknown function; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm and bud; interacts with Spa2p; YFL016C is not an essential gene |
||
| YOR346W | 0.10 |
REV1
|
Deoxycytidyl transferase, forms a complex with the subunits of DNA polymerase zeta, Rev3p and Rev7p; involved in repair of abasic sites in damaged DNA |
|
| YLL056C | 0.10 |
Putative protein of unknown function, transcription is activated by paralogous transcription factors Yrm1p and Yrr1p along with genes involved in pleiotropic drug resistance (PDR) phenomenon; YLL056C is not an essential gene |
||
| YPR065W | 0.10 |
ROX1
|
Heme-dependent repressor of hypoxic genes; contains an HMG domain that is responsible for DNA bending activity |
|
| YGR251W | 0.10 |
Putative protein of unknown function; deletion mutant has defects in pre-rRNA processing; green fluorescent protein (GFP)-fusion protein localizes to both the nucleus and the nucleolus; YGR251W is an essential gene |
||
| YFL021W | 0.10 |
GAT1
|
Transcriptional activator of genes involved in nitrogen catabolite repression; contains a GATA-1-type zinc finger DNA-binding motif; activity and localization regulated by nitrogen limitation and Ure2p |
|
| YER054C | 0.10 |
GIP2
|
Putative regulatory subunit of the protein phosphatase Glc7p, involved in glycogen metabolism; contains a conserved motif (GVNK motif) that is also found in Gac1p, Pig1p, and Pig2p |
|
| YNL187W | 0.10 |
Putative protein of unknown function; contains a consensus nuclear export signal (NES) sequence similar to the consensus sequence recognized by Crm1p; may interact with or be fuctionally redundant with Prp40p |
||
| YPR075C | 0.10 |
OPY2
|
Integral membrane protein that functions in the signaling branch of the high-osmolarity glycerol (HOG) pathway; interacts with Ste50p; overproduction blocks cell cycle arrest in the presence of mating pheromone |
|
| YDR313C | 0.10 |
PIB1
|
RING-type ubiquitin ligase of the endosomal and vacuolar membranes, binds phosphatidylinositol(3)-phosphate; contains a FYVE finger domain |
|
| YAR069C | 0.10 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YJR115W | 0.10 |
Putative protein of unknown function |
||
| YDL130W-A | 0.10 |
STF1
|
Protein involved in regulation of the mitochondrial F1F0-ATP synthase; Stf1p and Stf2p may act as stabilizing factors that enhance inhibitory action of the Inh1p protein |
|
| YLR312C | 0.09 |
QNQ1
|
Protein of unknown function; null mutant quiescent and non-quiescent cells exhibit reduced reproductive capacity |
|
| YBR076C-A | 0.09 |
Dubious open reading frame unlikely to encode a protein; partially overlaps verified gene ECM8; identified by fungal homology and RT-PCR |
||
| YLR296W | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YGR144W | 0.09 |
THI4
|
Thiazole synthase, catalyzes formation of the thiazole moiety of thiamin pyrophosphate; required for thiamine biosynthesis and for mitochondrial genome stability |
|
| YPR036W-A | 0.09 |
Protein of unknown function; transcription is regulated by Pdr1p |
||
| YOR192C | 0.09 |
THI72
|
Transporter of thiamine or related compound; shares sequence similarity with Thi7p |
|
| YBL100C | 0.09 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; almost completely overlaps the 5' end of ATP1 |
||
| YJR160C | 0.09 |
MPH3
|
Alpha-glucoside permease, transports maltose, maltotriose, alpha-methylglucoside, and turanose; identical to Mph2p; encoded in a subtelomeric position in a region likely to have undergone duplication |
|
| YOL082W | 0.09 |
ATG19
|
Receptor protein for the cytoplasm-to-vacuole targeting (Cvt) pathway; recognizes cargo proteins aminopeptidase I (Lap4p) and alpha-mannosidase (Ams1p) and delivers them to the preautophagosomal structure for packaging into Cvt vesicles |
|
| YJL133C-A | 0.09 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
||
| YDL182W | 0.09 |
LYS20
|
Homocitrate synthase isozyme, catalyzes the condensation of acetyl-CoA and alpha-ketoglutarate to form homocitrate, which is the first step in the lysine biosynthesis pathway; highly similar to the other isozyme, Lys21p |
|
| YKL166C | 0.09 |
TPK3
|
cAMP-dependent protein kinase catalytic subunit; promotes vegetative growth in response to nutrients via the Ras-cAMP signaling pathway; inhibited by regulatory subunit Bcy1p in the absence of cAMP; partially redundant with Tpk1p and Tpk2p |
|
| YGR180C | 0.09 |
RNR4
|
Ribonucleotide-diphosphate reductase (RNR), small subunit; the RNR complex catalyzes the rate-limiting step in dNTP synthesis and is regulated by DNA replication and DNA damage checkpoint pathways via localization of the small subunits |
|
| YMR175W | 0.09 |
SIP18
|
Protein of unknown function whose expression is induced by osmotic stress |
|
| YBR269C | 0.09 |
FMP21
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YDR042C | 0.09 |
Putative protein of unknown function; expression is increased in ssu72-ts69 mutant |
||
| YDR528W | 0.09 |
HLR1
|
Protein involved in regulation of cell wall composition and integrity and response to osmotic stress; overproduction suppresses a lysis sensitive PKC mutation; similar to Lre1p, which functions antagonistically to protein kinase A |
|
| YMR090W | 0.09 |
Putative protein of unknown function with similarity to DTDP-glucose 4,6-dehydratases; green fluorescent protein (GFP)-fusion protein localizes to the cytoplasm; YMR090W is not an essential gene |
||
| YLR236C | 0.08 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNR054C | 0.08 |
ESF2
|
Essential nucleolar protein involved in pre-18S rRNA processing; binds to RNA and stimulates ATPase activity of Dbp8; involved in assembly of the small subunit (SSU) processome |
|
| YML047C | 0.08 |
PRM6
|
Pheromone-regulated protein, predicted to have 2 transmembrane segments; regulated by Ste12p during mating |
|
| YML071C | 0.08 |
COG8
|
Component of the conserved oligomeric Golgi complex (Cog1p through Cog8p), a cytosolic tethering complex that functions in protein trafficking to mediate fusion of transport vesicles to Golgi compartments |
|
| YMR070W | 0.08 |
MOT3
|
Nuclear transcription factor with two Cys2-His2 zinc fingers; involved in repression of a subset of hypoxic genes by Rox1p, repression of several DAN/TIR genes during aerobic growth, and repression of ergosterol biosynthetic genes |
|
| YBR190W | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on experimental and comparative sequence data; partially overlaps the verified ribosomal protein gene RPL21A/YBR191W |
||
| YNL161W | 0.08 |
CBK1
|
Serine/threonine protein kinase that regulates cell morphogenesis pathways; involved in cell wall biosynthesis, apical growth, proper mating projection morphology, bipolar bud site selection in diploid cells, and cell separation |
|
| YDR299W | 0.08 |
BFR2
|
Essential protein possibly involved in secretion; multicopy suppressor of sensitivity to Brefeldin A |
|
| YMR320W | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YFL011W | 0.08 |
HXT10
|
Putative hexose transporter, expressed at low levels and expression is repressed by glucose |
|
| YDR155C | 0.08 |
CPR1
|
Cytoplasmic peptidyl-prolyl cis-trans isomerase (cyclophilin), catalyzes the cis-trans isomerization of peptide bonds N-terminal to proline residues; binds the drug cyclosporin A |
|
| YDL138W | 0.08 |
RGT2
|
Plasma membrane glucose receptor, highly similar to Snf3p; both Rgt2p and Snf3p serve as transmembrane glucose sensors generating an intracellular signal that induces expression of glucose transporter (HXT) genes |
|
| YPR076W | 0.08 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YNL014W | 0.08 |
HEF3
|
Translational elongation factor EF-3; paralog of YEF3 and member of the ABC superfamily; stimulates EF-1 alpha-dependent binding of aminoacyl-tRNA by the ribosome; normally expressed in zinc deficient cells |
|
| YJR048W | 0.08 |
CYC1
|
Cytochrome c, isoform 1; electron carrier of the mitochondrial intermembrane space that transfers electrons from ubiquinone-cytochrome c oxidoreductase to cytochrome c oxidase during cellular respiration |
|
| YER014C-A | 0.08 |
BUD25
|
Protein involved in bud-site selection; diploid mutants display a random budding pattern instead of the wild-type bipolar pattern |
|
| YOR229W | 0.08 |
WTM2
|
Transcriptional modulator involved in regulation of meiosis, silencing, and expression of RNR genes; involved in response to replication stress; contains WD repeats |
|
| YMR057C | 0.08 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps verified ORF AAC1 |
||
| YJR104C | 0.08 |
SOD1
|
Cytosolic superoxide dismutase; some mutations are analogous to those that cause ALS (amyotrophic lateral sclerosis) in humans |
|
| YDR542W | 0.08 |
PAU10
|
Hypothetical protein |
|
| YGR243W | 0.08 |
FMP43
|
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies |
|
| YML082W | 0.08 |
Putative protein predicted to have carbon-sulfur lyase activity; green fluorescent protein (GFP)-fusion protein localizes to the nucleus and the cytoplasm; YNML082W is not an essential gene |
||
| YER076W-A | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the uncharacterized ORF YER076C |
||
| YIL100C-A | 0.07 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YGL191W | 0.07 |
COX13
|
Subunit VIa of cytochrome c oxidase, which is the terminal member of the mitochondrial inner membrane electron transport chain; not essential for cytochrome c oxidase activity but may modulate activity in response to ATP |
|
| YER045C | 0.07 |
ACA1
|
Basic leucine zipper (bZIP) transcription factor of the ATF/CREB family, may regulate transcription of genes involved in utilization of non-optimal carbon sources |
|
| YLR193C | 0.07 |
UPS1
|
Mitochondrial intermembrane space protein that regulates alternative processing and sorting of Mgm1p and other proteins; required for normal mitochondrial morphology; ortholog of human PRELI |
|
| YOL026C | 0.07 |
MIM1
|
Mitochondrial outer membrane protein, required for assembly of the translocase of the outer membrane (TOM) complex and thereby for mitochondrial protein import; N terminus is exposed to the cytosol: transmembrane segment is highly conserved |
|
| YLR125W | 0.07 |
Putative protein of unknown function; mutant has decreased Ty3 transposition; YLR125W is not an essential gene |
||
| YKL096W | 0.07 |
CWP1
|
Cell wall mannoprotein, linked to a beta-1,3- and beta-1,6-glucan heteropolymer through a phosphodiester bond; involved in cell wall organization |
|
| YFL014W | 0.07 |
HSP12
|
Plasma membrane localized protein that protects membranes from desiccation; induced by heat shock, oxidative stress, osmostress, stationary phase entry, glucose depletion, oleate and alcohol; regulated by the HOG and Ras-Pka pathways |
|
| YPL036W | 0.07 |
PMA2
|
Plasma membrane H+-ATPase, isoform of Pma1p, involved in pumping protons out of the cell; regulator of cytoplasmic pH and plasma membrane potential |
|
| YOL156W | 0.07 |
HXT11
|
Putative hexose transporter that is nearly identical to Hxt9p, has similarity to major facilitator superfamily (MFS) transporters and is involved in pleiotropic drug resistance |
|
| YPL265W | 0.07 |
DIP5
|
Dicarboxylic amino acid permease, mediates high-affinity and high-capacity transport of L-glutamate and L-aspartate; also a transporter for Gln, Asn, Ser, Ala, and Gly |
|
| YPL054W | 0.07 |
LEE1
|
Zinc-finger protein of unknown function |
|
| YCR097W-A | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; identified by homology to a hemiascomycetous yeast protein |
||
| YMR172W | 0.07 |
HOT1
|
Transcription factor required for the transient induction of glycerol biosynthetic genes GPD1 and GPP2 in response to high osmolarity; targets Hog1p to osmostress responsive promoters; has similarity to Msn1p and Gcr1p |
|
| YKL109W | 0.07 |
HAP4
|
Subunit of the heme-activated, glucose-repressed Hap2p/3p/4p/5p CCAAT-binding complex, a transcriptional activator and global regulator of respiratory gene expression; provides the principal activation function of the complex |
|
| YMR096W | 0.07 |
SNZ1
|
Protein involved in vitamin B6 biosynthesis; member of a stationary phase-induced gene family; coregulated with SNO1; interacts with Sno1p and with Yhr198p, perhaps as a multiprotein complex containing other Snz and Sno proteins |
|
| YER076C | 0.07 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; analysis of HA-tagged protein suggests a membrane localization |
||
| YEL071W | 0.07 |
DLD3
|
D-lactate dehydrogenase, part of the retrograde regulon which consists of genes whose expression is stimulated by damage to mitochondria and reduced in cells grown with glutamate as the sole nitrogen source, located in the cytoplasm |
|
| YLR380W | 0.07 |
CSR1
|
Phosphatidylinositol transfer protein with a potential role in regulating lipid and fatty acid metabolism under heme-depleted conditions; interacts specifically with thioredoxin peroxidase; may have a role in oxidative stress resistance |
|
| YDR227W | 0.07 |
SIR4
|
Silent information regulator that, together with SIR2 and SIR3, is involved in assembly of silent chromatin domains at telomeres and the silent mating-type loci; potentially phosphorylated by Cdc28p; some alleles of SIR4 prolong lifespan |
|
| YPR169W-A | 0.07 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps two other dubious ORFs: YPR170C and YPR170W-B |
||
| YMR135W-A | 0.07 |
Dubious open reading frame unlikely to encode a functional protein, based on available experimental and comparative sequence data |
||
| YDL087C | 0.07 |
LUC7
|
Essential protein associated with the U1 snRNP complex; splicing factor involved in recognition of 5' splice site; contains two zinc finger motifs; N-terminal zinc finger binds pre-mRNA |
|
| YBR082C | 0.07 |
UBC4
|
Ubiquitin-conjugating enzyme (E2), mediates degradation of short-lived and abnormal proteins; interacts with E3-CaM in ubiquitinating calmodulin; interacts with many SCF ubiquitin protein ligases; component of the cellular stress response |
|
| YCL069W | 0.06 |
VBA3
|
Permease of basic amino acids in the vacuolar membrane |
|
| YML061C | 0.06 |
PIF1
|
DNA helicase involved in telomere formation and elongation; acts as a catalytic inhibitor of telomerase; also plays a role in repair and recombination of mitochondrial DNA |
|
| YMR172C-A | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data |
||
| YDL086W | 0.06 |
Putative protein of unknown function; the authentic, non-tagged protein is detected in highly purified mitochondria in high-throughput studies; YDL086W is not an essential gene |
||
| YDR171W | 0.06 |
HSP42
|
Small heat shock protein (sHSP) with chaperone activity; forms barrel-shaped oligomers that suppress unfolded protein aggregation; involved in cytoskeleton reorganization after heat shock |
|
| YPR096C | 0.06 |
Protein of unknown function that may interact with ribosomes, based on co-purification experiments |
||
| YGR294W | 0.06 |
PAU12
|
Hypothetical protein |
|
| YGR009C | 0.06 |
SEC9
|
t-SNARE protein important for fusion of secretory vesicles with the plasma membrane; similar to but not functionally redundant with Spo20p; SNAP-25 homolog |
|
| YEL045C | 0.06 |
Dubious open reading frame unlikely to encode a protein based on available experimental and comparative sequence data; deletion gives MMS sensitivity, growth defect under alkaline conditions, less than optimal growth upon citric acid stress |
||
| YBR230C | 0.06 |
OM14
|
Integral mitochondrial outer membrane protein; abundance is decreased in cells grown in glucose relative to other carbon sources; appears to contain 3 alpha-helical transmembrane segments; ORF encodes a 97-basepair intron |
|
| YOR055W | 0.06 |
Dubious open reading frame unlikely to encode a functional protein; based on available experimental and comparative sequence data |
||
| YML090W | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; partially overlaps the dubious ORF YML089C; exhibits growth defect on a non-fermentable (respiratory) carbon source |
||
| YBR081C | 0.06 |
SPT7
|
Subunit of the SAGA transcriptional regulatory complex, involved in proper assembly of the complex; also present as a C-terminally truncated form in the SLIK/SALSA transcriptional regulatory complex |
|
| YAR050W | 0.06 |
FLO1
|
Lectin-like protein involved in flocculation, cell wall protein that binds to mannose chains on the surface of other cells, confers floc-forming ability that is chymotrypsin sensitive and heat resistant; similar to Flo5p |
|
| YFR033C | 0.06 |
QCR6
|
Subunit 6 of the ubiquinol cytochrome-c reductase complex, which is a component of the mitochondrial inner membrane electron transport chain; highly acidic protein; required for maturation of cytochrome c1 |
|
| YPR013C | 0.06 |
Putative zinc finger protein; YPR013C is not an essential gene |
||
| YLR007W | 0.06 |
NSE1
|
Essential subunit of the Mms21-Smc5-Smc6 complex; nuclear protein required for DNA repair and growth; has a nonstructural role in the maintenance of chromosomes |
|
| YKL161C | 0.06 |
Protein kinase implicated in the Slt2p mitogen-activated (MAP) kinase signaling pathway; associates with Rlm1p |
||
| YDL011C | 0.06 |
Dubious open reading frame unlikely to encode a protein, based on available experimental and comparative sequence data; overlaps the uncharacterized ORF YDL010W |
||
| YOL110W | 0.06 |
SHR5
|
Subunit of a palmitoyltransferase, composed of Shr5p and Erf2p, that adds a palmitoyl lipid moiety to heterolipidated substrates such as Ras1p and Ras2p through a thioester linkage; palmitoylation is required for Ras2p membrane localization |
Network of associatons between targets according to the STRING database.
First level regulatory network of SKN7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0000949 | aromatic amino acid family catabolic process to alcohol via Ehrlich pathway(GO:0000949) |
| 0.1 | 0.5 | GO:0061416 | regulation of transcription from RNA polymerase II promoter in response to salt stress(GO:0061416) |
| 0.1 | 0.3 | GO:1900434 | filamentous growth of a population of unicellular organisms in response to starvation(GO:0036170) regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900434) positive regulation of filamentous growth of a population of unicellular organisms in response to starvation(GO:1900436) |
| 0.1 | 0.2 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.1 | 0.2 | GO:0043388 | positive regulation of DNA binding(GO:0043388) regulation of DNA binding(GO:0051101) |
| 0.1 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.5 | GO:0009746 | response to hexose(GO:0009746) response to glucose(GO:0009749) response to monosaccharide(GO:0034284) |
| 0.1 | 0.3 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.2 | GO:0071474 | cellular hyperosmotic response(GO:0071474) |
| 0.0 | 0.1 | GO:0072353 | age-dependent response to reactive oxygen species(GO:0001315) cellular age-dependent response to reactive oxygen species(GO:0072353) |
| 0.0 | 0.1 | GO:0019676 | ammonia assimilation cycle(GO:0019676) |
| 0.0 | 0.3 | GO:0000128 | flocculation(GO:0000128) flocculation via cell wall protein-carbohydrate interaction(GO:0000501) |
| 0.0 | 0.1 | GO:0045981 | positive regulation of nucleotide metabolic process(GO:0045981) positive regulation of purine nucleotide metabolic process(GO:1900544) |
| 0.0 | 0.1 | GO:0019401 | glycerol biosynthetic process(GO:0006114) alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.2 | GO:0046348 | aminoglycan catabolic process(GO:0006026) chitin catabolic process(GO:0006032) amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0051469 | vesicle fusion with vacuole(GO:0051469) |
| 0.0 | 0.1 | GO:2000058 | regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:2000058) |
| 0.0 | 0.1 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.2 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.4 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.1 | GO:0034389 | lipid particle organization(GO:0034389) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) |
| 0.0 | 0.2 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.5 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of deoxyribonuclease activity(GO:0032077) positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.1 | GO:0009415 | response to water(GO:0009415) cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.2 | GO:0070873 | regulation of glycogen biosynthetic process(GO:0005979) regulation of glycogen metabolic process(GO:0070873) |
| 0.0 | 0.1 | GO:0010674 | negative regulation of transcription from RNA polymerase II promoter involved in meiotic cell cycle(GO:0010674) |
| 0.0 | 0.2 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.1 | GO:0097201 | negative regulation of transcription from RNA polymerase II promoter in response to stress(GO:0097201) |
| 0.0 | 0.1 | GO:0031384 | regulation of initiation of mating projection growth(GO:0031384) |
| 0.0 | 0.4 | GO:0019740 | nitrogen utilization(GO:0019740) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.1 | GO:0015847 | putrescine transport(GO:0015847) |
| 0.0 | 0.2 | GO:0000196 | MAPK cascade involved in cell wall organization or biogenesis(GO:0000196) |
| 0.0 | 0.1 | GO:0001304 | progressive alteration of chromatin involved in replicative cell aging(GO:0001304) negative regulation of chromatin silencing involved in replicative cell aging(GO:0001308) |
| 0.0 | 0.1 | GO:0018063 | protein-heme linkage(GO:0017003) protein-tetrapyrrole linkage(GO:0017006) cytochrome c-heme linkage(GO:0018063) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.0 | GO:0015740 | C4-dicarboxylate transport(GO:0015740) |
| 0.0 | 0.3 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0052547 | negative regulation of peptidase activity(GO:0010466) regulation of peptidase activity(GO:0052547) |
| 0.0 | 0.1 | GO:1904357 | negative regulation of telomere maintenance via telomerase(GO:0032211) negative regulation of telomere maintenance via telomere lengthening(GO:1904357) |
| 0.0 | 0.2 | GO:0009651 | response to salt stress(GO:0009651) |
| 0.0 | 0.1 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.1 | GO:0022610 | cell adhesion(GO:0007155) biological adhesion(GO:0022610) |
| 0.0 | 0.0 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.2 | GO:0007121 | bipolar cellular bud site selection(GO:0007121) |
| 0.0 | 0.1 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0046184 | pyridoxal phosphate metabolic process(GO:0042822) pyridoxal phosphate biosynthetic process(GO:0042823) aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.0 | GO:0036095 | positive regulation of invasive growth in response to glucose limitation by positive regulation of transcription from RNA polymerase II promoter(GO:0036095) positive regulation of invasive growth in response to glucose limitation(GO:2000219) |
| 0.0 | 0.0 | GO:0033866 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0070987 | error-free translesion synthesis(GO:0070987) |
| 0.0 | 0.0 | GO:0010039 | response to iron ion(GO:0010039) regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) cellular response to iron ion(GO:0071281) |
| 0.0 | 0.1 | GO:0071709 | ascospore-type prospore membrane assembly(GO:0032120) membrane biogenesis(GO:0044091) membrane assembly(GO:0071709) |
| 0.0 | 0.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0006279 | premeiotic DNA replication(GO:0006279) |
| 0.0 | 0.1 | GO:0046777 | protein autophosphorylation(GO:0046777) |
| 0.0 | 0.1 | GO:0000114 | obsolete regulation of transcription involved in G1 phase of mitotic cell cycle(GO:0000114) |
| 0.0 | 0.3 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process from tryptophan(GO:0034354) 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0009228 | thiamine biosynthetic process(GO:0009228) thiamine-containing compound biosynthetic process(GO:0042724) |
| 0.0 | 0.1 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.0 | GO:0023021 | termination of signal transduction(GO:0023021) termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.0 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.0 | GO:0006075 | (1->3)-beta-D-glucan biosynthetic process(GO:0006075) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0019626 | propionate metabolic process(GO:0019541) propionate catabolic process(GO:0019543) short-chain fatty acid catabolic process(GO:0019626) propionate catabolic process, 2-methylcitrate cycle(GO:0019629) |
| 0.0 | 0.0 | GO:0032102 | negative regulation of macroautophagy(GO:0016242) negative regulation of response to external stimulus(GO:0032102) negative regulation of response to extracellular stimulus(GO:0032105) negative regulation of response to nutrient levels(GO:0032108) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031422 | RecQ helicase-Topo III complex(GO:0031422) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0044462 | cell wall part(GO:0044426) external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0005756 | mitochondrial proton-transporting ATP synthase, central stalk(GO:0005756) proton-transporting ATP synthase, central stalk(GO:0045269) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0000112 | nucleotide-excision repair factor 3 complex(GO:0000112) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.0 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0005784 | Sec61 translocon complex(GO:0005784) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0045259 | mitochondrial proton-transporting ATP synthase complex(GO:0005753) proton-transporting ATP synthase complex(GO:0045259) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0000133 | polarisome(GO:0000133) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.0 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0031307 | intrinsic component of mitochondrial outer membrane(GO:0031306) integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0034967 | Set3 complex(GO:0034967) |
| 0.0 | 0.0 | GO:0000148 | 1,3-beta-D-glucan synthase complex(GO:0000148) |
| 0.0 | 0.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0000974 | Prp19 complex(GO:0000974) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.1 | 0.5 | GO:0004737 | pyruvate decarboxylase activity(GO:0004737) |
| 0.1 | 0.5 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.1 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.3 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.1 | GO:0042947 | alpha-glucoside transmembrane transporter activity(GO:0015151) glucoside transmembrane transporter activity(GO:0042947) |
| 0.0 | 0.1 | GO:0004691 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0016208 | AMP binding(GO:0016208) |
| 0.0 | 0.0 | GO:0016880 | acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.2 | GO:0005354 | galactose transmembrane transporter activity(GO:0005354) |
| 0.0 | 0.4 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.2 | GO:0015205 | nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0004458 | D-lactate dehydrogenase (cytochrome) activity(GO:0004458) |
| 0.0 | 0.1 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups(GO:0016725) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0050839 | cell adhesion molecule binding(GO:0050839) |
| 0.0 | 0.3 | GO:0020037 | heme binding(GO:0020037) tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.1 | GO:0033549 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0008198 | ferrous iron binding(GO:0008198) |
| 0.0 | 0.1 | GO:0015116 | secondary active sulfate transmembrane transporter activity(GO:0008271) sulfate transmembrane transporter activity(GO:0015116) |
| 0.0 | 0.1 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) peptidase inhibitor activity(GO:0030414) |
| 0.0 | 0.2 | GO:0008599 | protein phosphatase type 1 regulator activity(GO:0008599) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) phosphatase inhibitor activity(GO:0019212) |
| 0.0 | 0.0 | GO:0003843 | 1,3-beta-D-glucan synthase activity(GO:0003843) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0044389 | ubiquitin protein ligase binding(GO:0031625) ubiquitin-like protein ligase binding(GO:0044389) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.0 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.1 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.2 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.0 | 0.1 | REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | Genes involved in Respiratory electron transport, ATP synthesis by chemiosmotic coupling, and heat production by uncoupling proteins. |
| 0.0 | 0.1 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |