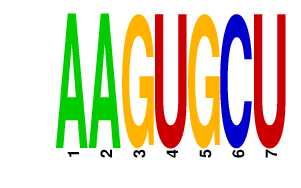
|
chr1_-_130488785
|
20.422
|
NM_009911
|
Cxcr4
|
chemokine (C-X-C motif) receptor 4
|
|
chr2_+_127161893
|
16.049
|
NM_010090
|
Dusp2
|
dual specificity phosphatase 2
|
|
chr2_+_179777187
|
13.624
|
NM_178750
|
Ss18l1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr10_+_96079634
|
13.126
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr12_+_72116953
|
12.521
|
NM_001081195
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr11_-_34597324
|
12.413
|
NM_033374
|
Dock2
|
dedicator of cyto-kinesis 2
|
|
chr4_+_135728188
|
12.127
|
NM_177733
|
E2f2
|
E2F transcription factor 2
|
|
chr14_+_56223363
|
11.625
|
NM_001159418
|
Irf9
|
interferon regulatory factor 9
|
|
chr14_+_56222821
|
11.397
|
NM_001159417
NM_008394
|
Irf9
|
interferon regulatory factor 9
|
|
chr3_-_30868320
|
11.179
|
NM_001165954
NM_001165955
NM_001165956
NM_153421
|
Phc3
|
polyhomeotic-like 3 (Drosophila)
|
|
chr13_+_41701584
|
11.033
|
NM_001163572
|
Tmem170b
|
transmembrane protein 170B
|
|
chr8_-_73316331
|
10.964
|
NM_199308
|
Mast3
|
microtubule associated serine/threonine kinase 3
|
|
chr4_+_114732112
|
10.668
|
NM_011527
|
Tal1
|
T-cell acute lymphocytic leukemia 1
|
|
chr11_-_90252228
|
9.311
|
NM_172563
|
Hlf
|
hepatic leukemia factor
|
|
chr12_+_85792371
|
9.006
|
NM_173756
|
Lin52
|
lin-52 homolog (C. elegans)
|
|
chr4_-_133443662
|
8.995
|
NM_009097
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1
|
|
chr4_+_135841981
|
8.819
|
NM_009557
|
Zfp46
|
zinc finger protein 46
|
|
chr2_-_131855722
|
8.760
|
NM_175445
|
Rassf2
|
Ras association (RalGDS/AF-6) domain family member 2
|
|
chr9_+_122026378
|
8.257
|
NM_001164572
NM_133741
|
Snrk
|
SNF related kinase
|
|
chr17_+_26852581
|
8.133
|
NM_029870
|
A930001N09Rik
|
RIKEN cDNA A930001N09 gene
|
|
chr9_-_44689356
|
8.014
|
NM_001081049
|
Mll1
|
myeloid/lymphoid or mixed-lineage leukemia 1
|
|
chr11_-_86807216
|
7.365
|
NM_001005341
|
Ypel2
|
yippee-like 2 (Drosophila)
|
|
chr1_+_179374783
|
7.304
|
NM_001012330
|
Zfp238
|
zinc finger protein 238
|
|
chr19_-_14672472
|
7.295
|
NM_011600
|
Tle4
|
transducin-like enhancer of split 4, homolog of Drosophila E(spl)
|
|
chr18_-_46358466
|
7.227
|
NM_001170855
|
Trim36
|
tripartite motif-containing 36
|
|
chr10_-_30522912
|
7.219
|
NM_172495
|
Ncoa7
|
nuclear receptor coactivator 7
|
|
chr17_+_25325344
|
7.168
|
NM_001197024
|
Unkl
|
unkempt-like (Drosophila)
|
|
chr5_-_44617844
|
7.100
|
NM_173764
|
Tapt1
|
transmembrane anterior posterior transformation 1
|
|
chr2_-_31939210
|
7.012
|
NM_175511
|
Fam78a
|
family with sequence similarity 78, member A
|
|
chr1_-_69732373
|
6.931
|
NM_011770
|
Ikzf2
|
IKAROS family zinc finger 2
|
|
chr5_+_37442095
|
6.868
|
NM_178394
|
Jakmip1
|
janus kinase and microtubule interacting protein 1
|
|
chr11_-_70501586
|
6.722
|
NM_001190376
NM_001190378
NM_001190379
NM_178116
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr18_+_51277551
|
6.663
|
NM_001081224
|
Prr16
|
proline rich 16
|
|
chr2_-_18919637
|
6.634
|
NM_008845
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr16_-_23988680
|
6.530
|
NM_009744
|
Bcl6
|
B-cell leukemia/lymphoma 6
|
|
chr10_+_40069113
|
6.420
|
NM_001168304
NM_198164
|
Cdk19
|
cyclin-dependent kinase 19
|
|
chr18_+_32322742
|
6.403
|
NM_011946
|
Map3k2
|
mitogen-activated protein kinase kinase kinase 2
|
|
chr3_+_60305108
|
6.396
|
NM_020007
|
Mbnl1
|
muscleblind-like 1 (Drosophila)
|
|
chr5_-_64360058
|
6.393
|
NM_145923
|
Rell1
|
RELT-like 1
|
|
chr11_+_60351159
|
6.314
|
NM_172943
|
Alkbh5
|
alkB, alkylation repair homolog 5 (E. coli)
|
|
chr6_-_90760116
|
6.205
|
NM_001134384
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr6_-_90666474
|
6.081
|
NM_001134383
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr13_-_112599181
|
6.012
|
NM_011945
|
Map3k1
|
mitogen-activated protein kinase kinase kinase 1
|
|
chr3_+_28162135
|
5.859
|
NM_001163007
NM_001163008
NM_001163009
NM_026910
|
Tnik
|
TRAF2 and NCK interacting kinase
|
|
chr5_-_138125817
|
5.852
|
NM_178162
|
Agfg2
|
ArfGAP with FG repeats 2
|
|
chr13_+_83643032
|
5.851
|
NM_001170537
NM_025282
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr6_-_116578970
|
5.819
|
NM_026057
|
Zfp422
|
zinc finger protein 422
|
|
chr10_+_42398721
|
5.775
|
NM_172416
|
Ostm1
|
osteopetrosis associated transmembrane protein 1
|
|
chr17_-_51951378
|
5.689
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr9_+_59598963
|
5.677
|
NM_173018
|
Myo9a
|
myosin IXa
|
|
chr5_+_139856534
|
5.659
|
NM_030258
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr17_-_51971946
|
5.625
|
NM_009122
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51965912
|
5.621
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51972514
|
5.590
|
NM_001163632
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr9_+_7764067
|
5.572
|
NM_133739
|
Tmem123
|
transmembrane protein 123
|
|
chr11_+_97524725
|
5.565
|
NM_139311
|
Mllt6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr2_+_173485260
|
5.565
|
NM_024436
|
Rab22a
|
RAB22A, member RAS oncogene family
|
|
chr17_+_25359526
|
5.555
|
NM_028789
|
Unkl
|
unkempt-like (Drosophila)
|
|
chr9_-_123760992
|
5.529
|
NM_001110253
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr17_-_31992708
|
5.526
|
NM_010831
|
Sik1
|
salt inducible kinase 1
|
|
chr4_-_128483355
|
5.502
|
NM_001081098
|
Zfp362
|
zinc finger protein 362
|
|
chr8_+_85387234
|
5.498
|
NM_177378
|
Rnf150
|
ring finger protein 150
|
|
chr6_-_28211600
|
5.492
|
NM_001081678
|
Zfp800
|
zinc finger protein 800
|
|
chr11_+_70376359
|
5.487
|
NM_001045959
NM_001045964
NM_016713
NM_176893
|
Mink1
|
misshapen-like kinase 1 (zebrafish)
|
|
chrX_-_48559008
|
5.330
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr6_+_92041408
|
5.308
|
NM_011630
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2
|
|
chr5_+_139865857
|
5.299
|
NM_001038703
|
Gpr146
|
G protein-coupled receptor 146
|
|
chrX_-_9046362
|
5.284
|
NM_007807
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr12_-_109241423
|
5.233
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr7_-_71083734
|
5.176
|
NM_021366
|
Klf13
|
Kruppel-like factor 13
|
|
chr11_-_4494815
|
5.078
|
NM_028860
|
Mtmr3
|
myotubularin related protein 3
|
|
chr16_-_92697567
|
5.058
|
NM_001111023
NM_009821
|
Runx1
|
runt related transcription factor 1
|
|
chr1_+_132613903
|
5.028
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr16_+_55973916
|
4.998
|
NM_173026
|
Zbtb11
|
zinc finger and BTB domain containing 11
|
|
chr12_-_119539909
|
4.997
|
NM_001166385
NM_009239
|
Sp4
|
trans-acting transcription factor 4
|
|
chr3_+_103718042
|
4.959
|
NM_172684
|
Rsbn1
|
rosbin, round spermatid basic protein 1
|
|
chr18_-_46372017
|
4.902
|
NM_178872
|
Trim36
|
tripartite motif-containing 36
|
|
chr3_+_88769733
|
4.874
|
NM_138679
|
Ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr15_+_100446018
|
4.826
|
NM_011873
|
Dazap2
|
DAZ associated protein 2
|
|
chr1_-_90598670
|
4.805
|
NM_177305
|
Arl4c
|
ADP-ribosylation factor-like 4C
|
|
chr4_+_105988790
|
4.798
|
NM_183225
|
Usp24
|
ubiquitin specific peptidase 24
|
|
chr11_-_5344795
|
4.738
|
NM_001080924
|
Znrf3
|
zinc and ring finger 3
|
|
chr14_-_46277816
|
4.716
|
NM_001039106
NM_001042719
NM_176845
|
Ddhd1
|
DDHD domain containing 1
|
|
chr2_-_37286367
|
4.710
|
NM_146253
|
Zbtb6
|
zinc finger and BTB domain containing 6
|
|
chr6_-_83391623
|
4.699
|
NM_183138
|
Tet3
|
tet oncogene family member 3
|
|
chr4_-_154010868
|
4.691
|
NM_001177995
NM_027504
|
Prdm16
|
PR domain containing 16
|
|
chr13_-_54150991
|
4.690
|
NM_010076
|
Drd1a
|
dopamine receptor D1A
|
|
chr11_+_117515548
|
4.680
|
NM_198022
|
Tnrc6c
|
trinucleotide repeat containing 6C
|
|
chr11_-_69761489
|
4.670
|
NM_009204
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr2_-_153067043
|
4.607
|
NM_018807
|
Plagl2
|
pleiomorphic adenoma gene-like 2
|
|
chr8_+_3393005
|
4.540
|
NM_133962
|
Arhgef18
|
rho/rac guanine nucleotide exchange factor (GEF) 18
|
|
chr13_+_112476313
|
4.533
|
NM_172593
|
Mier3
|
mesoderm induction early response 1, family member 3
|
|
chr6_+_122770201
|
4.504
|
NM_021899
|
Igfbp5
Foxj2
|
insulin-like growth factor binding protein 5
forkhead box J2
|
|
chr17_-_43803963
|
4.466
|
NM_028711
|
Slc25a27
|
solute carrier family 25, member 27
|
|
chr1_+_133904162
|
4.458
|
NM_007923
|
Elk4
|
ELK4, member of ETS oncogene family
|
|
chr6_+_91937103
|
4.403
|
NM_172731
|
Fgd5
|
FYVE, RhoGEF and PH domain containing 5
|
|
chr19_-_4397075
|
4.364
|
NM_001001984
|
Kdm2a
|
lysine (K)-specific demethylase 2A
|
|
chr11_+_69579376
|
4.358
|
NM_029348
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr10_-_30375572
|
4.343
|
NM_001111267
|
Ncoa7
|
nuclear receptor coactivator 7
|
|
chr6_-_39370351
|
4.318
|
NM_018810
|
Mkrn1
|
makorin, ring finger protein, 1
|
|
chr1_+_179375951
|
4.287
|
NM_013915
|
Zfp238
|
zinc finger protein 238
|
|
chr11_-_103128675
|
4.267
|
NM_016896
|
Map3k14
|
mitogen-activated protein kinase kinase kinase 14
|
|
chr2_+_127951773
|
4.191
|
NM_009754
NM_207680
NM_207681
|
Bcl2l11
|
BCL2-like 11 (apoptosis facilitator)
|
|
chr1_+_66515003
|
4.183
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr13_+_43493729
|
4.074
|
NM_023554
|
Nol7
|
nucleolar protein 7
|
|
chr10_+_127896244
|
4.042
|
NM_001114096
NM_001114097
NM_198160
|
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr11_-_88922061
|
4.040
|
NM_019505
|
Dgke
|
diacylglycerol kinase, epsilon
|
|
chr16_-_32079317
|
3.998
|
NM_177326
|
Pak2
|
p21 protein (Cdc42/Rac)-activated kinase 2
|
|
chr4_-_45024999
|
3.960
|
NM_001163283
NM_001163284
NM_173399
|
Zbtb5
|
zinc finger and BTB domain containing 5
|
|
chr4_-_44181148
|
3.952
|
NM_001038993
|
Rnf38
|
ring finger protein 38
|
|
chr2_+_22924561
|
3.939
|
NM_001102437
NM_001102438
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5
|
|
chr5_-_25004598
|
3.906
|
NM_001081383
|
Mll3
|
myeloid/lymphoid or mixed-lineage leukemia 3
|
|
chr19_-_47766512
|
3.901
|
NM_007732
|
Col17a1
|
collagen, type XVII, alpha 1
|
|
chr17_+_27110101
|
3.860
|
NM_001005916
|
Zbtb9
|
zinc finger and BTB domain containing 9
|
|
chr13_-_8870975
|
3.732
|
NM_001039388
|
Wdr37
|
WD repeat domain 37
|
|
chr3_+_58810899
|
3.713
|
NM_177855
|
Med12l
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast)-like
|
|
chr8_+_23970006
|
3.704
|
NM_001081149
|
Myst3
|
MYST histone acetyltransferase (monocytic leukemia) 3
|
|
chr15_+_80541675
|
3.619
|
NM_144812
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chrX_+_126284271
|
3.612
|
NM_172493
|
Diap2
|
diaphanous homolog 2 (Drosophila)
|
|
chr13_-_8870231
|
3.608
|
NM_001039389
NM_172445
|
Wdr37
|
WD repeat domain 37
|
|
chr1_-_34899878
|
3.607
|
NM_001160235
NM_001160236
NM_174997
|
Fam168b
|
family with sequence similarity 168, member B
|
|
chr2_+_32390780
|
3.604
|
NM_153560
|
Fam102a
|
family with sequence similarity 102, member A
|
|
chr4_-_126145660
|
3.581
|
NM_153403
|
Eif2c1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr4_-_131978279
|
3.566
|
NM_001161797
NM_175306
|
Phactr4
|
phosphatase and actin regulator 4
|
|
chr2_+_75670230
|
3.556
|
NM_172666
|
Agps
|
alkylglycerone phosphate synthase
|
|
chr9_+_57385317
|
3.552
|
NM_001166364
NM_175273
|
2310046O06Rik
|
RIKEN cDNA 2310046O06 gene
|
|
chr5_+_148242018
|
3.534
|
NM_028291
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr19_+_40905768
|
3.529
|
NM_172839
|
Ccnj
|
cyclin J
|
|
chr9_-_123760686
|
3.477
|
NM_148925
|
Fyco1
|
FYVE and coiled-coil domain containing 1
|
|
chr14_+_58691522
|
3.465
|
NM_013518
|
Fgf9
|
fibroblast growth factor 9
|
|
chr2_-_33323966
|
3.461
|
NM_001025594
NM_027947
|
Zbtb43
|
zinc finger and BTB domain containing 43
|
|
chr9_-_42280256
|
3.441
|
NM_173038
|
Tbcel
|
tubulin folding cofactor E-like
|
|
chr14_+_49065971
|
3.428
|
NM_172600
|
6720456H20Rik
|
RIKEN cDNA 6720456H20 gene
|
|
chr3_+_95462640
|
3.421
|
NM_008562
|
Mcl1
|
myeloid cell leukemia sequence 1
|
|
chr7_-_74517624
|
3.419
|
NM_001033713
|
Mef2a
|
myocyte enhancer factor 2A
|
|
chr14_+_104049453
|
3.410
|
NM_198014
|
Slain1
|
SLAIN motif family, member 1
|
|
chr9_-_51976215
|
3.399
|
NM_001162921
|
Zc3h12c
|
zinc finger CCCH type containing 12C
|
|
chr2_-_7317824
|
3.399
|
NM_001110231
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr2_+_22923653
|
3.369
|
NM_001102436
NM_028793
|
Acbd5
|
acyl-Coenzyme A binding domain containing 5
|
|
chrX_+_165233030
|
3.348
|
NM_009707
|
Arhgap6
|
Rho GTPase activating protein 6
|
|
chr17_-_88464582
|
3.317
|
NM_001081034
|
Fbxo11
|
F-box protein 11
|
|
chr5_-_138705172
|
3.295
|
NM_153161
|
BC037034
|
cDNA sequence BC037034
|
|
chrX_-_152057864
|
3.204
|
NM_001093750
|
Ptchd1
|
patched domain containing 1
|
|
chr11_+_95198329
|
3.178
|
NM_172543
|
Fam117a
|
family with sequence similarity 117, memberA
|
|
chr2_-_144096253
|
3.135
|
NM_024225
|
Snx5
|
sorting nexin 5
|
|
chr2_-_104656852
|
3.097
|
NM_001123327
|
Qser1
|
glutamine and serine rich 1
|
|
chrX_-_159326220
|
3.086
|
NM_025932
|
Syap1
|
synapse associated protein 1
|
|
chr9_-_62994774
|
3.044
|
NM_172446
NM_001163755
NM_001163757
|
Skor1
|
SKI family transcriptional corepressor 1
|
|
chr15_+_41662472
|
3.030
|
NM_001130164
|
Oxr1
3110047M12Rik
|
oxidation resistance 1
RIKEN cDNA 3110047M12 gene
|
|
chr19_+_3767420
|
3.028
|
NM_001167884
NM_001167885
NM_001167886
NM_001167887
NM_001167888
NM_001167889
NM_144871
|
Suv420h1
|
suppressor of variegation 4-20 homolog 1 (Drosophila)
|
|
chr2_+_118727248
|
3.026
|
NM_001045523
|
Bahd1
|
bromo adjacent homology domain containing 1
|
|
chr7_+_104728398
|
3.010
|
NM_001081267
|
Rsf1
|
remodeling and spacing factor 1
|
|
chr2_-_166888514
|
2.993
|
NM_001033196
|
Znfx1
|
zinc finger, NFX1-type containing 1
|
|
chr2_+_4480799
|
2.991
|
NM_001177844
|
Frmd4a
|
FERM domain containing 4A
|
|
chr11_-_51813857
|
2.986
|
NM_009458
|
Ube2b
|
ubiquitin-conjugating enzyme E2B, RAD6 homology (S. cerevisiae)
|
|
chr5_+_28643698
|
2.982
|
NM_028234
|
Rbm33
|
RNA binding motif protein 33
|
|
chr11_-_70832737
|
2.957
|
NM_033562
|
Derl2
|
Der1-like domain family, member 2
|
|
chr8_+_108583449
|
2.934
|
NM_010901
|
Nfatc3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3
|
|
chr12_-_80397239
|
2.929
|
NM_001008550
|
Zfyve26
|
zinc finger, FYVE domain containing 26
|
|
chr5_-_9161670
|
2.924
|
NM_001110327
NM_011806
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1
|
|
chrX_+_50265390
|
2.912
|
NM_027642
|
Phf6
|
PHD finger protein 6
|
|
chr13_-_64254466
|
2.889
|
NM_175494
|
Zfp367
|
zinc finger protein 367
|
|
chr14_+_37868015
|
2.851
|
NM_146245
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1
|
|
chrX_+_90531783
|
2.848
|
NM_007492
|
Arx
|
aristaless related homeobox
|
|
chr5_-_96590773
|
2.846
|
NM_178854
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like
|
|
chr19_+_40969176
|
2.822
|
NM_028319
|
E130314M14Rik
Zfp518a
|
RIKEN cDNA E130314M14 gene
zinc finger protein 518A
|
|
chr16_-_4559511
|
2.781
|
NM_031182
|
Tcfap4
|
transcription factor AP4
|
|
chr7_-_30290470
|
2.762
|
NM_001081028
|
Sipa1l3
|
signal-induced proliferation-associated 1 like 3
|
|
chr6_-_113991945
|
2.719
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr2_-_136213509
|
2.712
|
NM_172858
|
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr9_+_121669121
|
2.688
|
NM_001166644
|
Zfp651
|
zinc finger protein 651
|
|
chr2_+_4322020
|
2.682
|
NM_001177843
|
Frmd4a
|
FERM domain containing 4A
|
|
chr12_+_4082573
|
2.636
|
NM_153082
|
Dnajc27
|
DnaJ (Hsp40) homolog, subfamily C, member 27
|
|
chr17_+_78599556
|
2.634
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr2_-_157030236
|
2.626
|
NM_001139516
NM_011249
|
Rbl1
|
retinoblastoma-like 1 (p107)
|
|
chr13_-_9764376
|
2.617
|
NM_144516
|
Zmynd11
|
zinc finger, MYND domain containing 11
|
|
chr16_-_92826307
|
2.590
|
NM_001111021
NM_001111022
|
Runx1
|
runt related transcription factor 1
|
|
chr2_+_4073908
|
2.589
|
NM_172475
|
Frmd4a
|
FERM domain containing 4A
|
|
chr2_+_49921978
|
2.587
|
NM_177139
|
Lypd6
|
LY6/PLAUR domain containing 6
|
|
chr11_+_98657082
|
2.585
|
NM_028722
|
Msl1
|
male-specific lethal 1 homolog (Drosophila)
|
|
chrX_+_165474542
|
2.568
|
NM_178754
|
Arhgap6
|
Rho GTPase activating protein 6
|
|
chr15_-_103170514
|
2.557
|
NM_013866
|
Zfp385a
|
zinc finger protein 385A
|
|
chr13_-_117814322
|
2.553
|
NM_001081009
|
Parp8
|
poly (ADP-ribose) polymerase family, member 8
|
|
chr4_-_44180323
|
2.551
|
NM_175201
|
Rnf38
|
ring finger protein 38
|
|
chr5_-_37380147
|
2.525
|
NM_011716
|
Wfs1
|
Wolfram syndrome 1 homolog (human)
|
|
chr16_-_98144201
|
2.491
|
NM_174847
|
C2cd2
|
C2 calcium-dependent domain containing 2
|
|
chr9_-_62996767
|
2.485
|
NM_001163758
|
Skor1
|
SKI family transcriptional corepressor 1
|
|
chr6_-_116143380
|
2.482
|
NM_177412
|
Tmcc1
|
transmembrane and coiled coil domains 1
|
|
chr6_+_114593104
|
2.465
|
NM_028835
|
Atg7
|
autophagy-related 7 (yeast)
|
|
chr12_-_73337707
|
2.442
|
NM_001007596
|
Rtn1
|
reticulon 1
|
|
chr5_-_144493077
|
2.432
|
NM_029749
|
Usp42
|
ubiquitin specific peptidase 42
|
|
chr4_+_59818521
|
2.430
|
NM_172468
|
Snx30
|
sorting nexin family member 30
|
|
chr12_+_54349663
|
2.389
|
NM_013780
|
Npas3
|
neuronal PAS domain protein 3
|
|
chr6_+_53989925
|
2.388
|
NM_001163640
|
Chn2
|
chimerin (chimaerin) 2
|
|
chr13_+_14156038
|
2.335
|
NM_194262
NM_198122
|
Arid4b
|
AT rich interactive domain 4B (RBP1-like)
|
|
chr15_+_80629020
|
2.328
|
NM_177124
|
Tnrc6b
|
trinucleotide repeat containing 6b
|
|
chr9_-_119488133
|
2.310
|
NM_021544
|
Scn5a
|
sodium channel, voltage-gated, type V, alpha
|
|
chr10_-_114238419
|
2.302
|
NM_146241
|
Trhde
|
TRH-degrading enzyme
|
|
chr2_-_6805937
|
2.286
|
NM_001110228
NM_001160293
NM_010160
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr10_+_13686174
|
2.268
|
NM_010437
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2
|