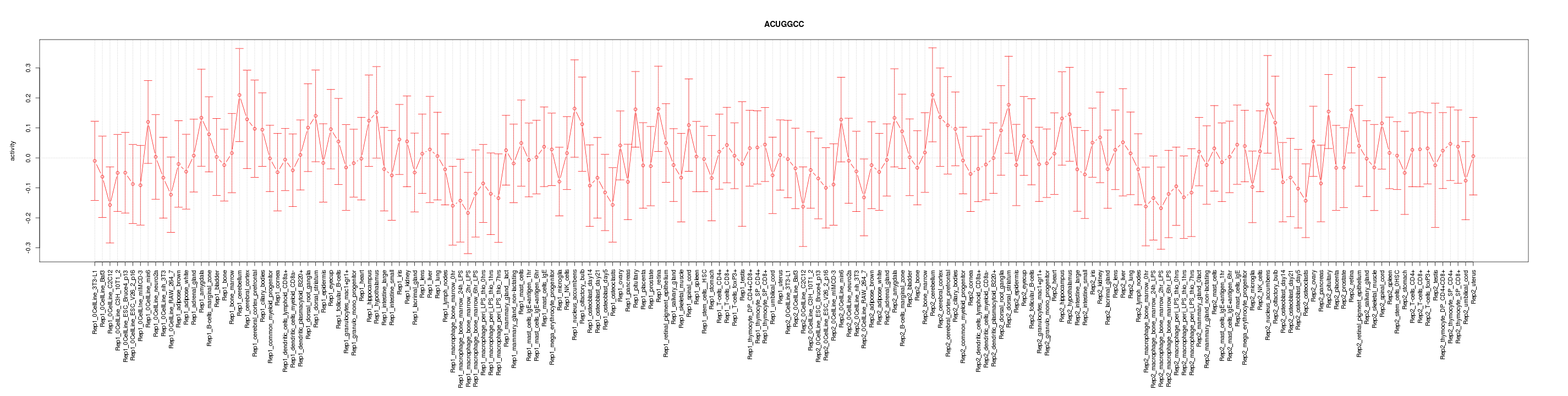
|
chr10_-_108447674
|
6.933
|
NM_009306
|
Syt1
|
synaptotagmin I
|
|
chr10_+_68996455
|
6.556
|
NM_146005
NM_170688
NM_170689
NM_170690
NM_170728
NM_170729
NM_170730
|
Ank3
|
ankyrin 3, epithelial
|
|
chr10_+_69388242
|
6.295
|
NM_009670
NM_170687
|
Ank3
|
ankyrin 3, epithelial
|
|
chrX_+_140127827
|
5.803
|
NM_001195046
NM_001195047
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr18_-_39078304
|
5.784
|
NM_010197
|
Fgf1
|
fibroblast growth factor 1
|
|
chr5_-_33196860
|
5.702
|
NM_194340
|
C330019G07Rik
|
RIKEN cDNA C330019G07 gene
|
|
chr18_-_15562126
|
5.379
|
NM_009700
|
Aqp4
|
aquaporin 4
|
|
chr5_-_103640272
|
5.315
|
NM_001081567
NM_009158
|
Mapk10
|
mitogen-activated protein kinase 10
|
|
chr4_+_15808409
|
4.927
|
NM_009788
|
Calb1
|
calbindin 1
|
|
chr18_+_65183180
|
4.865
|
NM_031881
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr5_+_75971011
|
4.726
|
NM_001122733
NM_021099
|
Kit
|
kit oncogene
|
|
chrX_+_139997806
|
4.708
|
NM_008778
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr12_-_15823590
|
4.494
|
NM_144551
|
Trib2
|
tribbles homolog 2 (Drosophila)
|
|
chrX_+_140100287
|
4.097
|
NM_001195049
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr17_+_8994520
|
3.960
|
NM_011866
|
Pde10a
|
phosphodiesterase 10A
|
|
chr1_-_72921438
|
3.946
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr4_-_43591721
|
3.905
|
NM_172692
|
Gba2
|
glucosidase beta 2
|
|
chr9_-_42913797
|
3.850
|
NM_027144
|
Arhgef12
|
Rho guanine nucleotide exchange factor (GEF) 12
|
|
chr4_-_151235861
|
3.717
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr4_+_13711928
|
3.611
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr18_+_65047408
|
3.443
|
NM_001114386
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr13_+_42804911
|
3.207
|
NM_001005740
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr5_-_51945054
|
3.175
|
NM_008904
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr7_+_104728398
|
3.066
|
NM_001081267
|
Rsf1
|
remodeling and spacing factor 1
|
|
chrX_+_139952975
|
2.836
|
NM_001195048
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr13_+_42775991
|
2.797
|
NM_198419
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr6_-_149050201
|
2.729
|
NM_177192
|
Dennd5b
|
DENN/MADD domain containing 5B
|
|
chr10_+_92623587
|
2.641
|
NM_146239
|
Cdk17
|
cyclin-dependent kinase 17
|
|
chr4_+_13670425
|
2.597
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr17_+_26852581
|
2.418
|
NM_029870
|
A930001N09Rik
|
RIKEN cDNA A930001N09 gene
|
|
chr13_+_43218207
|
2.374
|
NM_001005748
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr15_-_99358387
|
2.191
|
NM_001038658
NM_028224
|
Faim2
|
Fas apoptotic inhibitory molecule 2
|
|
chr9_+_32503600
|
2.177
|
NM_001038642
NM_011808
|
Ets1
|
E26 avian leukemia oncogene 1, 5' domain
|
|
chr1_-_58642994
|
2.158
|
NM_172513
|
Fam126b
|
family with sequence similarity 126, member B
|
|
chr1_+_174271338
|
2.120
|
NM_001039484
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr9_-_79641213
|
2.117
|
NM_133718
|
Tmem30a
|
transmembrane protein 30A
|
|
chr19_-_50752974
|
2.099
|
NM_021377
|
Sorcs1
|
VPS10 domain receptor protein SORCS 1
|
|
chr6_+_47194393
|
2.090
|
NM_025771
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr6_+_45010040
|
2.089
|
NM_001004357
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr13_-_52626065
|
2.053
|
NM_001024474
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr19_-_28057437
|
1.908
|
NM_001166414
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr4_+_40669478
|
1.815
|
NM_008298
|
Dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr12_+_39506844
|
1.815
|
NM_007960
|
Etv1
|
ets variant gene 1
|
|
chr15_+_81767188
|
1.800
|
NM_145473
|
Csdc2
|
cold shock domain containing C2, RNA binding
|
|
chr2_-_134469806
|
1.789
|
NM_029148
|
Tmx4
|
thioredoxin-related transmembrane protein 4
|
|
chr1_+_43155550
|
1.775
|
NM_133818
|
AI597479
|
expressed sequence AI597479
|
|
chr12_-_91976586
|
1.755
|
NM_010050
|
Dio2
|
deiodinase, iodothyronine, type II
|
|
chr11_+_105898183
|
1.709
|
NM_027946
|
Dcaf7
|
DDB1 and CUL4 associated factor 7
|
|
chr4_+_40669908
|
1.692
|
NM_001164671
NM_001164672
|
Dnaja1
|
DnaJ (Hsp40) homolog, subfamily A, member 1
|
|
chr15_-_79762947
|
1.676
|
NM_144811
|
Cbx7
|
chromobox homolog 7
|
|
chr18_-_75857207
|
1.583
|
NM_201354
|
Ctif
|
CBP80/20-dependent translation initiation factor
|
|
chr9_+_106073255
|
1.511
|
NM_029896
|
Wdr82
|
WD repeat domain containing 82
|
|
chr2_+_121782185
|
1.479
|
NM_212450
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr1_-_188529867
|
1.478
|
NM_009367
|
Tgfb2
|
transforming growth factor, beta 2
|
|
chr1_-_77511653
|
1.432
|
NM_007936
|
Epha4
|
Eph receptor A4
|
|
chr2_-_104250490
|
1.409
|
NM_001033347
|
D430041D05Rik
|
RIKEN cDNA D430041D05 gene
|
|
chr12_-_45311020
|
1.402
|
NM_013760
|
Dnajb9
|
DnaJ (Hsp40) homolog, subfamily B, member 9
|
|
chr3_-_146444377
|
1.306
|
NM_001164200
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr8_-_67373715
|
1.283
|
NM_178633
|
Klhl2
|
kelch-like 2, Mayven (Drosophila)
|
|
chr11_-_94183188
|
1.246
|
NM_026313
|
Luc7l3
|
LUC7-like 3 (S. cerevisiae)
|
|
chr3_-_95032589
|
1.132
|
NM_019914
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr3_+_151873407
|
1.124
|
NM_057172
|
Fubp1
|
far upstream element (FUSE) binding protein 1
|
|
chr1_+_65233222
|
1.105
|
NM_011086
|
Pikfyve
|
phosphoinositide kinase, FYVE finger containing
|
|
chr16_+_33684551
|
1.091
|
NM_175256
|
Heg1
|
HEG homolog 1 (zebrafish)
|
|
chr12_+_39510297
|
0.989
|
NM_001163154
|
Etv1
|
ets variant gene 1
|
|
chrX_-_12339096
|
0.960
|
NM_001048208
NM_012005
|
Med14
|
mediator complex subunit 14
|
|
chr4_-_45024999
|
0.939
|
NM_001163283
NM_001163284
NM_173399
|
Zbtb5
|
zinc finger and BTB domain containing 5
|
|
chr12_+_88019649
|
0.889
|
NM_177354
|
Vash1
|
vasohibin 1
|
|
chr19_-_28085534
|
0.840
|
NM_011265
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression)
|
|
chr2_+_109757803
|
0.801
|
NM_172671
|
Lgr4
|
leucine-rich repeat-containing G protein-coupled receptor 4
|
|
chr5_+_139865857
|
0.785
|
NM_001038703
|
Gpr146
|
G protein-coupled receptor 146
|
|
chr2_+_173902521
|
0.772
|
NM_001102423
NM_001102424
NM_001102425
NM_172675
|
Stx16
|
syntaxin 16
|
|
chr3_-_146433544
|
0.763
|
NM_001164199
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr12_-_77811533
|
0.669
|
NM_013675
|
Spnb1
|
spectrin beta 1
|
|
chr9_+_113840051
|
0.616
|
NM_001083319
NM_013699
|
Ubp1
|
upstream binding protein 1
|
|
chr4_-_141346551
|
0.489
|
NM_172338
|
Dnajc16
|
DnaJ (Hsp40) homolog, subfamily C, member 16
|
|
chr5_-_107718503
|
0.463
|
NM_011578
|
Tgfbr3
|
transforming growth factor, beta receptor III
|
|
chr11_-_98407281
|
0.383
|
NM_011771
|
Ikzf3
|
IKAROS family zinc finger 3
|
|
chr2_+_132088974
|
0.361
|
NM_138651
|
Cds2
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2
|
|
chr4_+_149493452
|
0.360
|
NM_019741
|
Slc2a5
|
solute carrier family 2 (facilitated glucose transporter), member 5
|
|
chr18_+_69505374
|
0.356
|
NM_001083967
|
Tcf4
|
transcription factor 4
|
|
chr4_+_125168074
|
0.340
|
NM_001081097
|
Grik3
|
glutamate receptor, ionotropic, kainate 3
|
|
chr13_-_34437042
|
0.325
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr2_+_30093236
|
0.276
|
NM_177725
|
Lrrc8a
|
leucine rich repeat containing 8A
|
|
chr5_-_124482223
|
0.247
|
NM_177876
|
Vps37b
|
vacuolar protein sorting 37B (yeast)
|
|
chr19_+_8816292
|
0.226
|
NM_019829
|
Stx5a
|
syntaxin 5A
|
|
chr3_-_146433200
|
0.210
|
NM_001164198
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr6_-_144157971
|
0.196
|
NM_001113559
NM_011444
|
Sox5
|
SRY-box containing gene 5
|
|
chr2_+_158620555
|
0.185
|
NM_145742
|
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chr9_-_44689356
|
0.135
|
NM_001081049
|
Mll1
|
myeloid/lymphoid or mixed-lineage leukemia 1
|
|
chr7_-_25372613
|
0.134
|
NM_001033205
|
Zfp575
|
zinc finger protein 575
|
|
chr2_-_160698705
|
0.116
|
NM_177263
|
Zhx3
|
zinc fingers and homeoboxes 3
|
|
chr8_-_97637241
|
0.114
|
NM_001145832
|
Kifc3
|
kinesin family member C3
|
|
chr13_-_94269616
|
0.102
|
NM_021310
|
Jmy
|
junction-mediating and regulatory protein
|
|
chr8_-_89269853
|
0.074
|
NM_009172
|
Siah1a
|
seven in absentia 1A
|
|
chr19_+_8815893
|
0.047
|
NM_001167799
|
Stx5a
|
syntaxin 5A
|
|
chr3_+_90058166
|
0.038
|
NM_028881
|
Crtc2
|
CREB regulated transcription coactivator 2
|
|
chr4_-_53172748
|
0.025
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|