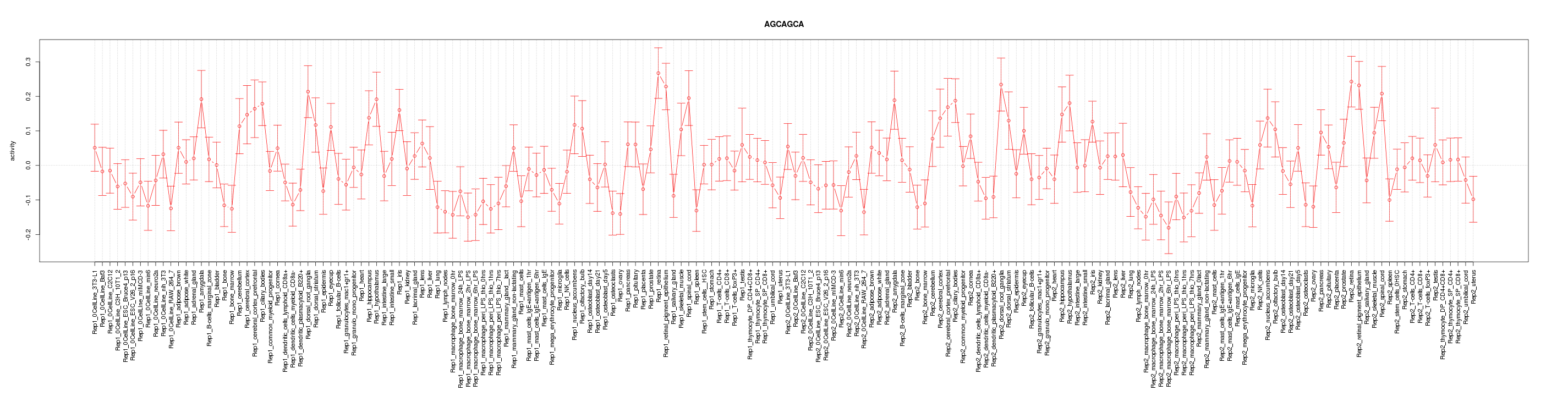
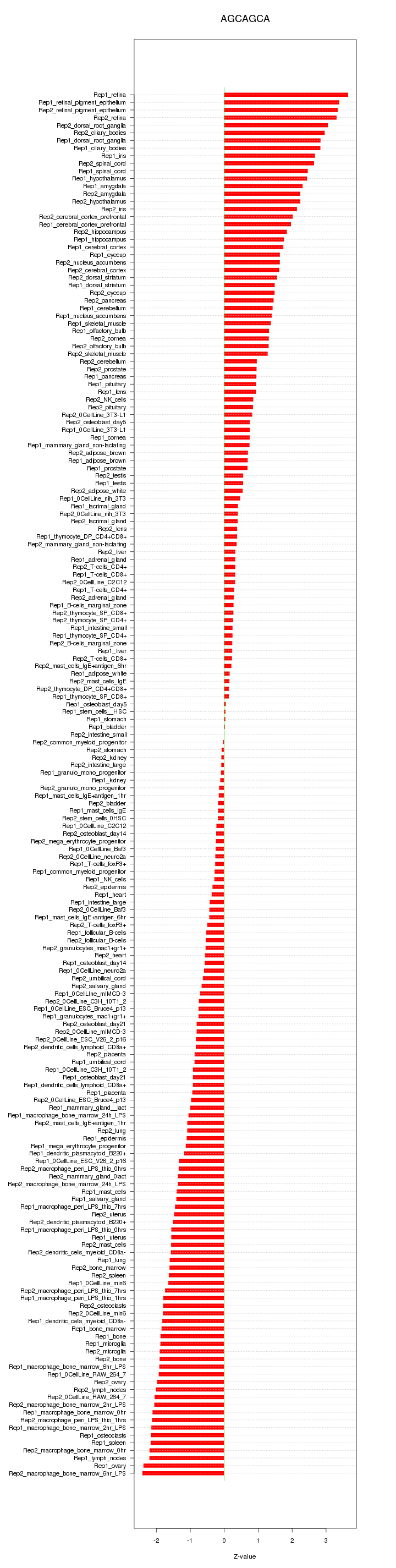
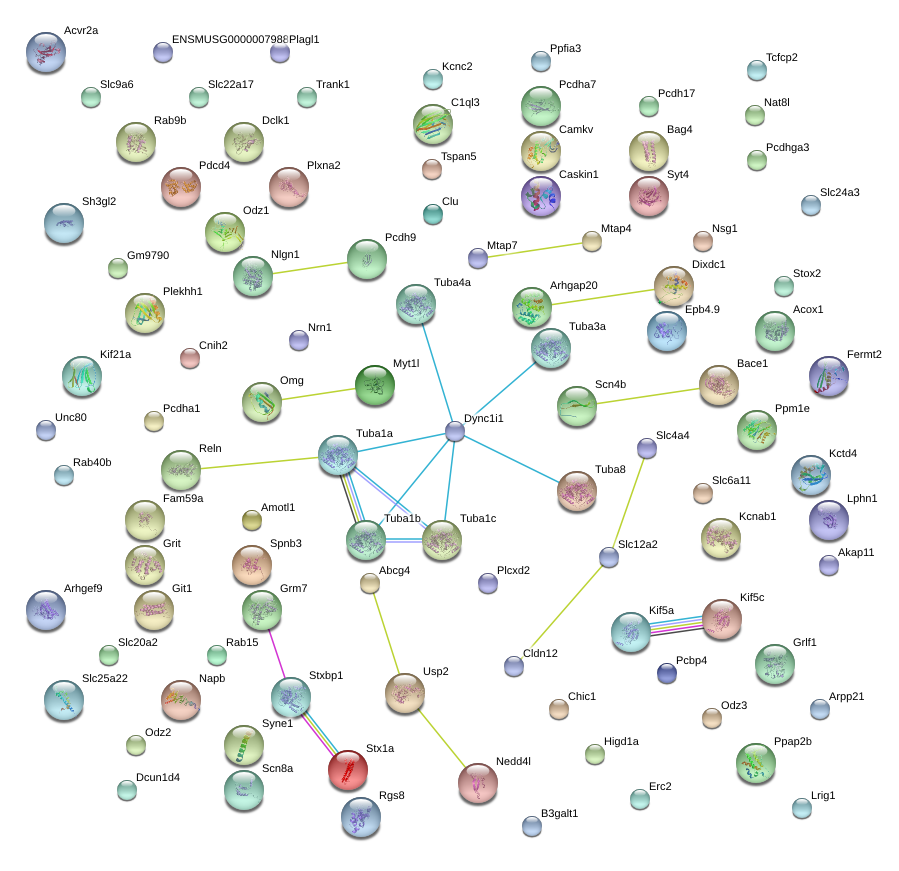
|
chr15_-_90880241
|
24.781
|
NM_001109040
NM_001109041
NM_001109042
NM_016705
|
Kif21a
|
kinesin family member 21A
|
|
chr1_+_66515003
|
23.493
|
NM_175510
|
Unc80
|
unc-80 homolog (C. elegans)
|
|
chr4_+_84851330
|
23.203
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr11_-_79317583
|
20.313
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr19_+_4711220
|
20.130
|
NM_021287
|
Spnb3
|
spectrin beta 3
|
|
chr2_+_49474775
|
19.917
|
NM_008449
|
Kif5c
|
kinesin family member 5C
|
|
chr18_-_31606908
|
17.623
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chr14_-_78936606
|
17.567
|
NM_001164503
|
Akap11
|
A kinase (PRKA) anchor protein 11
|
|
chr18_+_37098858
|
17.469
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr14_-_94287950
|
17.348
|
NM_001081377
|
Pcdh9
|
protocadherin 9
|
|
chr18_+_37179802
|
17.220
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chr9_+_32031833
|
17.137
|
NM_177379
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr11_-_36757743
|
16.975
|
NM_011856
|
Odz2
|
odd Oz/ten-m homolog 2 (Drosophila)
|
|
chr14_+_28435613
|
16.937
|
NM_177814
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr18_+_37089837
|
16.901
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr15_+_100701068
|
16.888
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr18_+_37120093
|
16.787
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr15_+_100766701
|
16.697
|
NM_011323
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr5_+_34338625
|
15.766
|
NM_001001985
|
Nat8l
|
N-acetyltransferase 8-like
|
|
chr18_+_37127284
|
15.758
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr14_-_46149660
|
15.272
|
NM_146054
|
Fermt2
|
fermitin family homolog 2 (Drosophila)
|
|
chr16_-_46010510
|
15.261
|
NM_001134480
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr9_+_51573434
|
14.908
|
NM_175535
|
Arhgap20
|
Rho GTPase activating protein 20
|
|
chr3_+_64913564
|
14.857
|
NM_010597
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr18_+_37105758
|
14.755
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr10_+_111708176
|
14.645
|
NM_001025581
|
Kcnc2
|
potassium voltage gated channel, Shaw-related subfamily, member 2
|
|
chr2_-_148558110
|
14.600
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr18_+_37157533
|
14.393
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chr2_+_145068346
|
14.211
|
NM_053195
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr18_+_37152024
|
14.131
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr18_+_37133455
|
14.105
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr8_-_48437701
|
14.053
|
NM_175162
|
Stox2
|
storkhead box 2
|
|
chr18_+_37303622
|
13.977
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr14_+_84843366
|
13.344
|
NM_001013753
|
Pcdh17
|
protocadherin 17
|
|
chr2_+_67955769
|
13.230
|
NM_020283
|
B3galt1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1
|
|
chr9_+_111214242
|
13.143
|
NM_001164659
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr8_+_86423968
|
13.065
|
NM_181039
|
Lphn1
|
latrophilin 1
|
|
chr5_-_5514787
|
12.929
|
NM_001193660
NM_001193659
NM_001193661
NM_022890
|
Cldn12
|
claudin 12
|
|
chr2_-_32702729
|
12.826
|
NM_009295
NM_001113569
|
Stxbp1
|
syntaxin binding protein 1
|
|
chr6_+_110595586
|
12.772
|
NM_177328
|
Grm7
|
glutamate receptor, metabotropic 7
|
|
chr8_-_48374755
|
12.328
|
NM_001114311
|
Stox2
|
storkhead box 2
|
|
chr12_+_30219769
|
12.317
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr9_-_121767026
|
12.122
|
NM_019814
|
Higd1a
|
HIG1 domain family, member 1A
|
|
chr8_-_26895633
|
11.978
|
NM_026121
|
Bag4
|
BCL2-associated athanogene 4
|
|
chr14_+_66587319
|
11.967
|
NM_013492
|
Clu
|
clusterin
|
|
chr1_+_155500166
|
11.843
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chr11_-_87172419
|
11.815
|
NM_177167
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing)
|
|
chrX_+_100551808
|
11.532
|
NM_009767
|
Chic1
|
cysteine-rich hydrophobic domain 1
|
|
chr10_+_19868725
|
11.438
|
NM_001198635
NM_008635
|
Mtap7
|
microtubule-associated protein 7
|
|
chr9_-_112134984
|
11.412
|
NM_001177616
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr9_+_45646611
|
11.282
|
NM_001145947
NM_011792
|
Bace1
|
beta-site APP cleaving enzyme 1
|
|
chr5_+_135499346
|
11.191
|
NM_016801
|
Stx1a
|
syntaxin 1A (brain)
|
|
chr10_+_5277477
|
11.149
|
NM_022027
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr9_+_31923720
|
11.146
|
NM_001195632
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr5_-_38550627
|
11.132
|
NM_010942
|
Nsg1
|
neuron specific gene family member 1
|
|
chr1_+_196446006
|
11.102
|
NM_008882
|
Plxna2
|
plexin A2
|
|
chrX_-_92361778
|
10.958
|
NM_001033329
|
Arhgef9
|
CDC42 guanine nucleotide exchange factor (GEF) 9
|
|
chr18_-_21458623
|
10.757
|
NM_001033445
|
Fam59a
|
family with sequence similarity 59, member A
|
|
chr6_+_5675638
|
10.752
|
NM_001191023
NM_001191025
NM_001191026
NM_001191027
NM_010063
|
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1
|
|
chr9_+_44947045
|
10.699
|
NM_001013390
|
Scn4b
|
sodium channel, type IV, beta
|
|
chr9_-_112088452
|
10.688
|
NM_001177618
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr5_-_21850490
|
10.666
|
NM_011261
|
Reln
|
reelin
|
|
chr2_-_12932442
|
10.575
|
NM_153155
|
C1ql3
|
C1q-like 3
|
|
chr9_+_43877511
|
10.526
|
NM_016808
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr14_+_76354807
|
10.523
|
NM_026214
|
Kctd4
|
potassium channel tetramerisation domain containing 4
|
|
chr9_+_107838164
|
10.464
|
NM_145621
|
Camkv
|
CaM kinase-like vesicle-associated
|
|
chr5_+_73872293
|
10.458
|
NM_001190734
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr9_+_43892984
|
10.456
|
NM_198091
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr9_+_43875103
|
10.443
|
NM_198092
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr6_-_94650118
|
10.282
|
NM_008377
|
Lrig1
|
leucine-rich repeats and immunoglobulin-like domains 1
|
|
chr9_+_109834266
|
10.255
|
NM_008633
|
Mtap4
|
microtubule-associated protein 4
|
|
chr3_+_55265654
|
10.182
|
NM_001111051
NM_001111052
NM_001195539
NM_001195540
|
Dclk1
|
doublecortin-like kinase 1
|
|
chr14_-_55531726
|
10.129
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr3_+_55046447
|
10.108
|
NM_001111053
NM_001195538
NM_019978
|
Dclk1
|
doublecortin-like kinase 1
|
|
chr8_-_49760037
|
10.102
|
NM_001145937
NM_011857
|
Odz3
|
odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr10_+_5151833
|
9.901
|
NM_001079686
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr10_-_126700387
|
9.838
|
NM_001039000
NM_008447
|
Kif5a
|
kinesin family member 5A
|
|
chr17_+_24625709
|
9.786
|
NM_027937
|
Caskin1
|
CASK interacting protein 1
|
|
chr9_+_106356125
|
9.740
|
NM_021567
|
Pcbp4
|
poly(rC) binding protein 4
|
|
chr7_-_17200285
|
9.724
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr6_+_114081227
|
9.559
|
NM_172890
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr12_+_30213224
|
9.349
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chrX_-_133403050
|
9.342
|
NM_176971
|
Rab9b
|
RAB9B, member RAS oncogene family
|
|
chr5_+_73882264
|
9.318
|
NM_001190733
NM_178896
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr3_-_26052228
|
9.262
|
NM_001163387
|
Nlgn1
|
neuroligin 1
|
|
chr5_+_89316245
|
9.220
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr11_+_77306825
|
9.198
|
NM_001004144
|
Git1
|
G protein-coupled receptor kinase-interactor 1
|
|
chr8_+_23587151
|
9.165
|
NM_011394
|
Slc20a2
|
solute carrier family 20, member 2
|
|
chr19_-_5098381
|
9.059
|
NM_009920
|
Cnih2
|
cornichon homolog 2 (Drosophila)
|
|
chr18_+_37112301
|
9.031
|
NM_001174154
NM_007766
|
Pcdha4-g
Pcdha4
|
protocadherin alpha 4-gamma
protocadherin alpha 4
|
|
chr10_+_12810593
|
8.844
|
NM_009538
|
Plagl1
|
pleiomorphic adenoma gene-like 1
|
|
chr9_-_44096170
|
8.716
|
NM_138955
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr3_-_26230791
|
8.647
|
NM_138666
|
Nlgn1
|
neuroligin 1
|
|
chr11_-_116060358
|
8.566
|
NM_015729
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl
|
|
chrX_+_53862956
|
8.562
|
NM_172780
|
Slc9a6
|
solute carrier family 9 (sodium/hydrogen exchanger), member 6
|
|
chr14_-_71029960
|
8.535
|
NM_013514
|
Epb4.9
|
erythrocyte protein band 4.9
|
|
chr19_+_53966720
|
8.343
|
NM_011050
|
Pdcd4
|
programmed cell death 4
|
|
chr2_+_48669628
|
8.230
|
NM_007396
|
Acvr2a
|
activin receptor IIA
|
|
chr18_+_58038278
|
8.218
|
NM_009194
|
Slc12a2
|
solute carrier family 12, member 2
|
|
chr3_+_138405156
|
8.218
|
NM_019571
|
Tspan5
|
tetraspanin 5
|
|
chr7_-_52622373
|
8.209
|
NM_029741
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr9_-_14419410
|
8.196
|
NM_001081395
|
Amotl1
|
angiomotin-like 1
|
|
chr9_-_50535949
|
8.184
|
NM_178118
|
Dixdc1
|
DIX domain containing 1
|
|
chr15_-_100382354
|
8.039
|
NM_033476
|
Tcfcp2
|
transcription factor CP2
|
|
chr15_-_98783909
|
8.028
|
NM_011653
|
Tuba1a
|
tubulin, alpha 1A
|
|
chr12_-_77923303
|
7.998
|
NM_134050
|
Rab15
|
RAB15, member RAS oncogene family
|
|
chr12_+_80130149
|
7.864
|
NM_181073
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chr4_+_104829951
|
7.860
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr18_+_65183180
|
7.806
|
NM_031881
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr11_-_121249511
|
7.803
|
NM_139147
|
Rab40b
|
Rab40b, member RAS oncogene family
|
|
chr13_-_36826245
|
7.784
|
NM_153529
|
Nrn1
|
neuritin 1
|
|
chr7_-_148623725
|
7.764
|
NM_026646
|
Slc25a22
|
solute carrier family 25 (mitochondrial carrier, glutamate), member 22
|
|
chr10_+_105907394
|
7.710
|
NM_177373
|
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2
|
|
chr2_+_109533719
|
7.611
|
NM_001048142
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr6_+_149357407
|
7.602
|
NM_001112796
NM_009753
|
Bicd1
|
bicaudal D homolog 1 (Drosophila)
|
|
chr11_-_119903061
|
7.594
|
NM_001198787
|
Aatk
|
apoptosis-associated tyrosine kinase
|
|
chr19_-_46647546
|
7.540
|
NM_019718
|
Arl3
|
ADP-ribosylation factor-like 3
|
|
chr4_+_43888373
|
7.447
|
NM_016678
|
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr4_+_80556574
|
7.414
|
NM_026821
|
D4Bwg0951e
|
DNA segment, Chr 4, Brigham & Women's Genetics 0951 expressed
|
|
chr4_+_151712739
|
7.331
|
NM_001081376
NM_029216
|
Chd5
|
chromodomain helicase DNA binding protein 5
|
|
chr2_-_136213509
|
7.288
|
NM_172858
|
Pak7
|
p21 protein (Cdc42/Rac)-activated kinase 7
|
|
chr11_+_87405628
|
7.281
|
NM_133215
|
Mtmr4
|
myotubularin related protein 4
|
|
chr10_-_78054639
|
7.277
|
NM_027875
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr4_-_75779379
|
7.262
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr6_-_113841356
|
7.261
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr4_-_147278927
|
7.169
|
NM_133201
|
Mfn2
|
mitofusin 2
|
|
chr9_+_21742717
|
7.144
|
NM_144935
|
BC018242
|
cDNA sequence BC018242
|
|
chr13_+_41345120
|
7.123
|
NM_001135577
|
BC024659
|
cDNA sequence BC024659
|
|
chr8_-_74195650
|
7.076
|
NM_001029873
|
Unc13a
|
unc-13 homolog A (C. elegans)
|
|
chr3_+_5218545
|
7.073
|
NM_030708
|
Zfhx4
|
zinc finger homeodomain 4
|
|
chr7_+_29477867
|
7.068
|
NM_207238
|
Fbxo27
|
F-box protein 27
|
|
chr8_+_97296066
|
6.923
|
NM_009142
|
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr4_-_76240202
|
6.874
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chrX_-_153929935
|
6.830
|
NM_009214
|
Sms
|
spermine synthase
|
|
chr3_+_84668066
|
6.812
|
NM_001177774
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr6_+_29718424
|
6.812
|
NM_001171000
NM_021414
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2
|
|
chr2_+_31101302
|
6.794
|
NM_019681
|
Ncs1
|
neuronal calcium sensor 1
|
|
chr15_-_100325669
|
6.782
|
NM_153407
|
Csrnp2
|
cysteine-serine-rich nuclear protein 2
|
|
chrY_-_623013
|
6.770
|
NM_012008
|
Ddx3y
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked
|
|
chr10_-_112365976
|
6.766
|
NM_001033474
|
Atxn7l3b
|
ataxin 7-like 3B
|
|
chr5_+_48374328
|
6.753
|
NM_178804
|
Slit2
|
slit homolog 2 (Drosophila)
|
|
chr2_+_149656518
|
6.750
|
NM_001085521
|
Tmem90b
|
transmembrane protein 90B
|
|
chr3_+_40549475
|
6.711
|
NM_011020
|
Hspa4l
|
heat shock protein 4 like
|
|
chr18_+_65739883
|
6.642
|
NM_207255
|
Zfp532
|
zinc finger protein 532
|
|
chr3_+_98186380
|
6.634
|
NM_172863
|
Zfp697
|
zinc finger protein 697
|
|
chr19_-_60019266
|
6.549
|
NM_001033172
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I)
|
|
chr5_+_118426774
|
6.519
|
NM_145564
|
Fbxo21
|
F-box protein 21
|
|
chr2_-_37502738
|
6.459
|
NM_009261
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr6_-_113991945
|
6.458
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_+_125165597
|
6.423
|
NM_001080557
NM_009496
|
Vamp1
|
vesicle-associated membrane protein 1
|
|
chr3_-_126646303
|
6.389
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr8_-_107995282
|
6.322
|
NM_026481
|
Tppp3
|
tubulin polymerization-promoting protein family member 3
|
|
chr4_-_148681792
|
6.310
|
NM_008441
NM_207682
|
Kif1b
|
kinesin family member 1B
|
|
chr10_+_21712008
|
6.164
|
NM_001161849
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr14_-_70999625
|
6.125
|
NM_194345
|
Fam160b2
|
family with sequence similarity 160, member B2
|
|
chr10_-_42894259
|
6.081
|
NM_175407
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr8_+_3515373
|
6.080
|
NM_001122818
NM_015801
|
Pnpla6
|
patatin-like phospholipase domain containing 6
|
|
chrX_-_131343759
|
6.061
|
NM_001166397
NM_001166398
NM_026139
|
Armcx2
|
armadillo repeat containing, X-linked 2
|
|
chr15_-_81791230
|
6.048
|
NM_013872
|
Pmm1
|
phosphomannomutase 1
|
|
chr18_+_65047408
|
5.992
|
NM_001114386
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like
|
|
chr19_-_60019520
|
5.865
|
NM_001164367
|
Rab11fip2
|
RAB11 family interacting protein 2 (class I)
|
|
chr18_+_23573908
|
5.797
|
NM_010087
NM_207650
|
Dtna
|
dystrobrevin alpha
|
|
chr17_+_78599556
|
5.779
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr9_+_108710676
|
5.735
|
NM_030729
|
Nckipsd
|
NCK interacting protein with SH3 domain
|
|
chr9_-_77392505
|
5.733
|
NM_001146048
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr2_+_109532592
|
5.664
|
NM_001048141
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr19_-_19185685
|
5.634
|
NM_001043354
|
Rorb
|
RAR-related orphan receptor beta
|
|
chr10_-_17954752
|
5.624
|
NM_144820
|
Ccdc28a
|
coiled-coil domain containing 28A
|
|
chr15_-_101966611
|
5.600
|
NM_001033277
|
Spryd3
|
SPRY domain containing 3
|
|
chr14_-_65716932
|
5.554
|
NM_018788
|
Extl3
|
exostoses (multiple)-like 3
|
|
chr5_+_91946651
|
5.539
|
NM_145562
|
Parm1
|
prostate androgen-regulated mucin-like protein 1
|
|
chr18_+_37249789
|
5.535
|
NM_001003671
|
Pcdhac1
|
protocadherin alpha subfamily C, 1
|
|
chr6_-_56747675
|
5.488
|
NM_145958
|
Kbtbd2
|
kelch repeat and BTB (POZ) domain containing 2
|
|
chr2_-_139892477
|
5.449
|
NM_175225
NM_001159640
NM_001159641
|
Tasp1
|
taspase, threonine aspartase 1
|
|
chr2_+_124436031
|
5.446
|
NM_172537
NM_199238
NM_199239
NM_199240
NM_199241
|
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D
|
|
chr8_+_42325093
|
5.446
|
NM_023662
|
Pcm1
|
pericentriolar material 1
|
|
chr19_-_14672472
|
5.440
|
NM_011600
|
Tle4
|
transducin-like enhancer of split 4, homolog of Drosophila E(spl)
|
|
chr10_+_21698470
|
5.388
|
NM_001161847
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr2_-_29979973
|
5.384
|
NM_178694
|
Zer1
|
zer-1 homolog (C. elegans)
|
|
chr2_-_151458256
|
5.239
|
NM_198214
|
Snph
|
syntaphilin
|
|
chr2_-_17652669
|
5.211
|
NM_028757
|
Nebl
|
nebulette
|
|
chr1_-_21952022
|
5.189
|
NM_001160139
NM_023872
|
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5
|
|
chr9_+_122714664
|
5.187
|
NM_001013405
|
D9Ertd402e
|
DNA segment, Chr 9, ERATO Doi 402, expressed
|
|
chr2_+_173563060
|
5.185
|
NM_019806
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C
|
|
chr4_-_45097427
|
5.163
|
NM_001024142
|
Fbxo10
|
F-box protein 10
|
|
chr5_+_122161617
|
5.162
|
NM_009125
|
Atxn2
|
ataxin 2
|
|
chr3_+_84619399
|
5.162
|
NM_001177773
|
Fbxw7
|
F-box and WD-40 domain protein 7
|
|
chr16_+_38089086
|
5.126
|
NM_019827
|
Gsk3b
|
glycogen synthase kinase 3 beta
|
|
chr6_+_8900018
|
5.098
|
NM_008751
|
Nxph1
|
neurexophilin 1
|
|
chr3_+_88769733
|
5.086
|
NM_138679
|
Ash1l
|
ash1 (absent, small, or homeotic)-like (Drosophila)
|
|
chr9_-_99336923
|
5.081
|
NM_008624
|
Mras
|
muscle and microspikes RAS
|
|
chr7_+_29478162
|
5.048
|
NM_001163702
|
Fbxo27
|
F-box protein 27
|
|
chrX_+_45265614
|
5.003
|
NM_177215
|
Ocrl
|
oculocerebrorenal syndrome of Lowe
|
|
chr2_-_160934743
|
4.967
|
NM_173368
|
Chd6
|
chromodomain helicase DNA binding protein 6
|
|
chr11_-_108205199
|
4.966
|
NM_011101
|
Prkca
|
protein kinase C, alpha
|
|
chr10_+_57718246
|
4.942
|
NM_027375
|
Gcc2
|
GRIP and coiled-coil domain containing 2
|
|
chr1_-_108656198
|
4.917
|
NM_027534
|
Kdsr
|
3-ketodihydrosphingosine reductase
|
|
chr3_+_114783882
|
4.913
|
NM_153157
|
Olfm3
|
olfactomedin 3
|
|
chr2_-_38920925
|
4.891
|
NM_029793
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1
|
|
chr8_+_74432504
|
4.884
|
NM_001166645
|
Zfp882
|
zinc finger protein 882
|