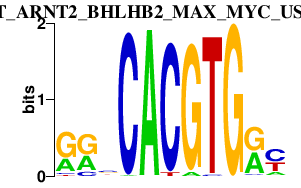
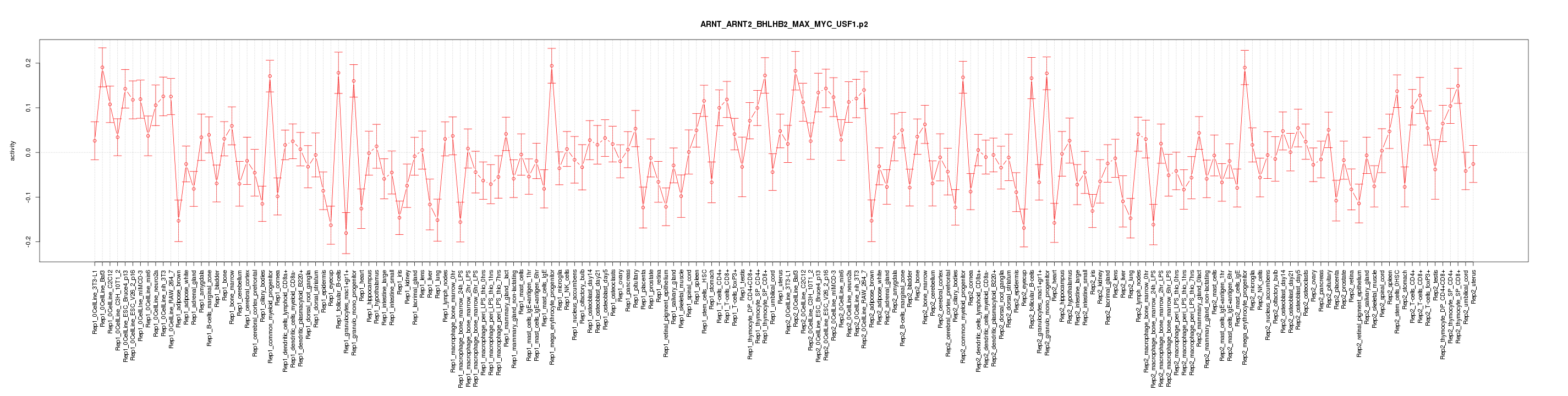
|
chr10_+_126500607
|
88.202
|
NM_009870
|
Cdk4
|
cyclin-dependent kinase 4
|
|
chr14_+_51544629
|
53.467
|
NM_009687
|
Apex1
|
apurinic/apyrimidinic endonuclease 1
|
|
chr9_+_21172314
|
41.889
|
NM_001042707
NM_001042708
NM_001042709
NM_010561
|
Ilf3
|
interleukin enhancer binding factor 3
|
|
chr19_-_4201541
|
41.200
|
NM_011237
|
Rad9
|
RAD9 homolog (S. pombe)
|
|
chr9_+_21216277
|
35.101
|
NM_021888
|
Qtrt1
|
queuine tRNA-ribosyltransferase 1
|
|
chr6_-_88848650
|
30.975
|
NM_008564
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae)
|
|
chr17_-_35137799
|
30.418
|
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5
|
|
chr1_+_59741800
|
30.387
|
NM_018868
|
Nop58
|
NOP58 ribonucleoprotein homolog (yeast)
|
|
chr11_+_69814046
|
30.196
|
NM_007888
|
Dvl2
|
dishevelled 2, dsh homolog (Drosophila)
|
|
chr4_+_147965596
|
26.175
|
NM_009272
|
Srm
|
spermidine synthase
|
|
chr5_-_138613028
|
26.148
|
NM_008568
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae)
|
|
chr4_+_134024234
|
25.763
|
NM_019641
|
Stmn1
|
stathmin 1
|
|
chr5_-_77380593
|
24.904
|
NM_172146
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr7_-_52722144
|
24.873
|
NM_007527
|
Bax
|
BCL2-associated X protein
|
|
chr17_+_35137995
|
23.831
|
|
Vars
|
valyl-tRNA synthetase
|
|
chr17_-_35137690
|
22.958
|
|
D17H6S56E-5
|
DNA segment, Chr 17, human D6S56E 5
|
|
chr15_+_59205752
|
22.681
|
NM_001164604
NM_026746
|
Nsmce2
|
non-SMC element 2 homolog (MMS21, S. cerevisiae)
|
|
chr3_+_89577436
|
22.201
|
NM_027315
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q (putative) 1
|
|
chrX_-_131076136
|
21.868
|
NM_013898
|
Timm8a1
|
translocase of inner mitochondrial membrane 8 homolog a1 (yeast)
|
|
chr17_-_53828596
|
21.756
|
NM_028232
|
Sgol1
|
shugoshin-like 1 (S. pombe)
|
|
chr14_-_51544484
|
21.524
|
|
Osgep
|
O-sialoglycoprotein endopeptidase
|
|
chr4_-_34830178
|
20.053
|
NM_013889
|
Zfp292
|
zinc finger protein 292
|
|
chr14_-_51544362
|
19.729
|
|
Osgep
|
O-sialoglycoprotein endopeptidase
|
|
chrX_+_45499599
|
18.973
|
NM_028773
|
Sash3
|
SAM and SH3 domain containing 3
|
|
chr16_+_35770488
|
18.694
|
|
Hspbap1
|
Hspb associated protein 1
|
|
chrX_+_149574541
|
18.665
|
|
Rragb
|
Ras-related GTP binding B
|
|
chr3_-_89856250
|
18.527
|
NM_001165983
NM_001165985
NM_001165987
NM_028475
NM_153489
NM_001165984
NM_001165986
NM_001165988
|
Ubap2l
|
ubiquitin associated protein 2-like
|
|
chr2_+_158620555
|
17.424
|
NM_145742
|
Dhx35
|
DEAH (Asp-Glu-Ala-His) box polypeptide 35
|
|
chrX_+_48194918
|
17.399
|
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene
|
|
chr10_+_43198945
|
17.397
|
NM_199028
|
Bend3
|
BEN domain containing 3
|
|
chrX_+_132804834
|
17.280
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr19_+_7049425
|
17.246
|
NM_008889
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chrX_+_149574498
|
17.133
|
NM_001004154
|
Rragb
|
Ras-related GTP binding B
|
|
chr11_-_93747039
|
17.063
|
NM_001013375
|
Utp18
|
UTP18, small subunit (SSU) processome component, homolog (yeast)
|
|
chr11_-_94515067
|
17.056
|
NM_177752
|
Eme1
|
essential meiotic endonuclease 1 homolog 1 (S. pombe)
|
|
chr8_+_120022149
|
16.794
|
NM_172285
|
Plcg2
|
phospholipase C, gamma 2
|
|
chr3_+_159158396
|
16.708
|
NM_001172092
NM_001172093
NM_029523
|
Depdc1a
|
DEP domain containing 1a
|
|
chrX_+_132804865
|
16.670
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr2_+_30141908
|
16.574
|
NM_198304
|
Nup188
|
nucleoporin 188
|
|
chr4_+_140258090
|
16.366
|
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr4_+_140257340
|
16.365
|
NM_173867
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr11_-_93746982
|
16.347
|
|
Utp18
|
UTP18, small subunit (SSU) processome component, homolog (yeast)
|
|
chr19_-_4615180
|
16.344
|
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr11_-_94515034
|
16.270
|
|
Eme1
|
essential meiotic endonuclease 1 homolog 1 (S. pombe)
|
|
chr17_+_29169612
|
16.232
|
|
Srsf3
|
serine/arginine-rich splicing factor 3
|
|
chr2_-_84510822
|
16.192
|
NM_001033166
NM_001037279
|
2700094K13Rik
|
RIKEN cDNA 2700094K13 gene
|
|
chr15_+_103070840
|
16.093
|
NM_001039129
NM_010447
|
Hnrnpa1
|
heterogeneous nuclear ribonucleoprotein A1
|
|
chr10_-_119913990
|
16.017
|
NM_010441
|
Hmga2
|
high mobility group AT-hook 2
|
|
chr17_-_33822327
|
15.858
|
NM_001109913
NM_029804
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr3_+_67386633
|
15.744
|
NM_025813
|
Mfsd1
|
major facilitator superfamily domain containing 1
|
|
chr17_+_35137851
|
15.182
|
NM_011690
|
Vars
|
valyl-tRNA synthetase
|
|
chr17_+_29169579
|
14.975
|
NM_013663
|
Srsf3
|
serine/arginine-rich splicing factor 3
|
|
chr4_-_48486214
|
14.951
|
NM_172304
|
Tex10
|
testis expressed gene 10
|
|
chr3_+_152059620
|
14.884
|
NM_198416
|
Zzz3
|
zinc finger, ZZ domain containing 3
|
|
chr11_+_68859125
|
14.807
|
NM_011496
|
Aurkb
|
aurora kinase B
|
|
chr18_+_56867378
|
14.716
|
NM_010721
|
Lmnb1
|
lamin B1
|
|
chr5_-_136646451
|
14.705
|
NM_178751
|
Orai2
|
ORAI calcium release-activated calcium modulator 2
|
|
chr17_+_29169665
|
14.650
|
|
Srsf3
|
serine/arginine-rich splicing factor 3
|
|
chr16_-_5204069
|
14.570
|
|
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chrX_+_132804857
|
14.449
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr16_-_5204078
|
14.370
|
NM_013796
|
Nagpa
|
N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase
|
|
chr11_-_117641895
|
14.349
|
NM_181321
|
Tmc6
|
transmembrane channel-like gene family 6
|
|
chr10_+_82092582
|
14.328
|
NM_011561
NM_172552
|
Tdg
Gm9855
|
thymine DNA glycosylase
thymine DNA glycosylase pseudogene
|
|
chr17_+_45701100
|
14.125
|
NM_028662
|
Slc35b2
|
solute carrier family 35, member B2
|
|
chr1_+_168309297
|
14.047
|
NM_030245
|
Tada1
|
transcriptional adaptor 1
|
|
chr16_+_35770431
|
13.801
|
NM_175111
|
Hspbap1
|
Hspb associated protein 1
|
|
chr13_+_21272294
|
13.602
|
|
Trim27
|
tripartite motif-containing 27
|
|
chr1_+_174430081
|
13.371
|
|
Tagln2
|
transgelin 2
|
|
chr11_-_84326869
|
13.341
|
NM_019816
|
Aatf
|
apoptosis antagonizing transcription factor
|
|
chr10_+_79479416
|
13.326
|
NM_001142701
|
Hmha1
|
histocompatibility (minor) HA-1
|
|
chr11_+_87569075
|
13.249
|
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene
|
|
chr11_-_102742230
|
13.249
|
NM_001109995
NM_011431
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2
|
|
chr1_-_60154406
|
13.006
|
|
Wdr12
|
WD repeat domain 12
|
|
chr17_+_46633703
|
12.951
|
NM_207161
|
BC048355
|
cDNA sequence BC048355
|
|
chr2_-_103601232
|
12.908
|
NM_153126
|
Nat10
|
N-acetyltransferase 10
|
|
chr3_-_101091814
|
12.877
|
NM_013486
|
Cd2
|
CD2 antigen
|
|
chr11_-_117641951
|
12.763
|
NM_145439
|
Tmc6
|
transmembrane channel-like gene family 6
|
|
chr17_+_71965554
|
12.761
|
NM_175639
|
Wdr43
|
WD repeat domain 43
|
|
chr19_-_4615460
|
12.754
|
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr4_+_132091462
|
12.693
|
NM_172400
|
Dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr19_-_10278231
|
12.648
|
NM_007999
|
Fen1
|
flap structure specific endonuclease 1
|
|
chr14_-_51544497
|
12.502
|
NM_133676
|
Osgep
|
O-sialoglycoprotein endopeptidase
|
|
chr3_+_138190223
|
12.437
|
|
Eif4e
|
eukaryotic translation initiation factor 4E
|
|
chr3_-_107889507
|
12.284
|
NM_028779
|
Ampd2
|
adenosine monophosphate deaminase 2
|
|
chr7_-_52241669
|
12.262
|
NM_019830
|
Prmt1
|
protein arginine N-methyltransferase 1
|
|
chr5_-_111209468
|
12.251
|
NM_001025561
NM_019700
NM_001025562
|
Pus1
|
pseudouridine synthase 1
|
|
chr5_-_114280434
|
12.233
|
NM_009151
|
Selplg
|
selectin, platelet (p-selectin) ligand
|
|
chr5_+_110715337
|
12.088
|
NM_011132
|
Pole
|
polymerase (DNA directed), epsilon
|
|
chr8_+_87213209
|
11.970
|
|
Trmt1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr4_-_133574655
|
11.927
|
NM_145833
|
Lin28a
|
lin-28 homolog A (C. elegans)
|
|
chr8_+_87213179
|
11.851
|
|
Trmt1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr17_-_32540473
|
11.829
|
|
Rasal3
|
RAS protein activator like 3
|
|
chr5_-_77380189
|
11.816
|
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr5_+_120566489
|
11.803
|
NM_028762
|
Rbm19
|
RNA binding motif protein 19
|
|
chr7_-_134404120
|
11.773
|
NM_023203
|
Dctpp1
|
dCTP pyrophosphatase 1
|
|
chr1_-_55144637
|
11.693
|
|
Hspd1
|
heat shock protein 1 (chaperonin)
|
|
chr2_+_75497282
|
11.608
|
NM_053263
NM_146130
NM_198090
|
Hnrnpa3
Gm6793
|
heterogeneous nuclear ribonucleoprotein A3
heterogeneous nuclear ribonucleoprotein A3 pseudogene
|
|
chrX_+_136991190
|
11.604
|
|
Prps1
|
phosphoribosyl pyrophosphate synthetase 1
|
|
chr1_-_55144687
|
11.534
|
NM_010477
|
Hspd1
|
heat shock protein 1 (chaperonin)
|
|
chr10_+_79317396
|
11.463
|
|
Ptbp1
|
polypyrimidine tract binding protein 1
|
|
chrX_+_132804791
|
11.388
|
NM_001110233
NM_001110234
NM_009750
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr7_+_104845148
|
11.324
|
NM_023671
|
Clns1a
|
chloride channel, nucleotide-sensitive, 1A
|
|
chr19_-_4615661
|
11.302
|
NM_153388
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr5_-_77380226
|
11.239
|
|
Ppat
|
phosphoribosyl pyrophosphate amidotransferase
|
|
chr10_+_79317316
|
11.209
|
NM_001077363
NM_008956
|
Ptbp1
|
polypyrimidine tract binding protein 1
|
|
chr13_+_69750927
|
11.144
|
|
Nsun2
|
NOL1/NOP2/Sun domain family member 2
|
|
chr4_+_88783273
|
11.136
|
NM_024433
|
Mtap
|
methylthioadenosine phosphorylase
|
|
chr2_+_130100145
|
11.102
|
NM_024193
|
Nop56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr16_+_45224449
|
11.093
|
NM_001037719
NM_177584
|
Btla
|
B and T lymphocyte associated
|
|
chr15_-_96473193
|
10.974
|
NM_001166458
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr4_-_41222121
|
10.921
|
NM_026872
|
Ubap2
|
ubiquitin-associated protein 2
|
|
chr1_+_55145132
|
10.821
|
|
Hspe1
|
heat shock protein 1 (chaperonin 10)
|
|
chr3_+_105762425
|
10.788
|
|
Wdr77
|
WD repeat domain 77
|
|
chr3_+_102862151
|
10.781
|
NM_010937
|
Nras
|
neuroblastoma ras oncogene
|
|
chr17_-_32540518
|
10.777
|
NM_178785
|
Rasal3
|
RAS protein activator like 3
|
|
chr4_+_122960059
|
10.769
|
NM_130881
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4
|
|
chr10_+_5914137
|
10.720
|
NM_025343
|
Rmnd1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae)
|
|
chr4_-_136604609
|
10.715
|
NM_198248
|
Zbtb40
|
zinc finger and BTB domain containing 40
|
|
chr4_-_147412872
|
10.603
|
NM_011929
|
Clcn6
|
chloride channel 6
|
|
chr8_+_87213145
|
10.602
|
NM_001164559
NM_001164560
NM_198020
|
Trmt1
|
TRM1 tRNA methyltransferase 1 homolog (S. cerevisiae)
|
|
chr8_+_90796301
|
10.583
|
NM_007406
|
Adcy7
|
adenylate cyclase 7
|
|
chr1_+_89223558
|
10.537
|
NM_146112
NM_001110212
|
Gigyf2
|
GRB10 interacting GYF protein 2
|
|
chr7_-_52384104
|
10.463
|
NM_009438
|
Rpl13a
|
ribosomal protein L13A
|
|
chr15_-_59205688
|
10.392
|
NM_153548
|
E430025E21Rik
|
RIKEN cDNA E430025E21 gene
|
|
chr8_+_126545374
|
10.328
|
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae)
|
|
chr6_-_38249209
|
10.283
|
NM_172467
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like
|
|
chr19_+_7049639
|
10.241
|
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B
|
|
chrX_+_156065141
|
10.203
|
NM_001135727
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr5_-_101227486
|
10.158
|
NM_001081107
|
Helq
|
helicase, POLQ-like
|
|
chr16_-_92826307
|
10.072
|
NM_001111021
NM_001111022
|
Runx1
|
runt related transcription factor 1
|
|
chr10_-_5913894
|
10.052
|
NM_024261
|
1700052N19Rik
|
RIKEN cDNA 1700052N19 gene
|
|
chr3_+_105762363
|
10.018
|
NM_027432
|
Wdr77
|
WD repeat domain 77
|
|
chr11_+_57822347
|
9.987
|
NM_028451
|
Larp1
|
La ribonucleoprotein domain family, member 1
|
|
chr17_-_25097542
|
9.954
|
NM_198937
|
Hn1l
|
hematological and neurological expressed 1-like
|
|
chr8_+_86239094
|
9.940
|
NM_197982
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39
|
|
chr4_+_132091510
|
9.895
|
|
Dnajc8
|
DnaJ (Hsp40) homolog, subfamily C, member 8
|
|
chr19_-_4615523
|
9.867
|
|
Lrfn4
|
leucine rich repeat and fibronectin type III domain containing 4
|
|
chr3_+_137527575
|
9.867
|
|
H2afz
|
H2A histone family, member Z
|
|
chr15_+_101904294
|
9.825
|
|
Eif4b
|
eukaryotic translation initiation factor 4B
|
|
chr17_+_45700882
|
9.796
|
|
Slc35b2
|
solute carrier family 35, member B2
|
|
chr2_-_164658954
|
9.756
|
NM_001082974
|
Neurl2
|
neuralized-like 2 (Drosophila)
|
|
chr1_-_127808790
|
9.741
|
NM_145100
|
Lypd1
|
Ly6/Plaur domain containing 1
|
|
chr7_-_36587975
|
9.739
|
NM_177739
|
Zfp507
E130304I02Rik
|
zinc finger protein 507
RIKEN cDNA E130304I02 gene
|
|
chr1_-_127808723
|
9.637
|
|
Lypd1
|
Ly6/Plaur domain containing 1
|
|
chr12_+_17551771
|
9.621
|
|
Odc1
|
ornithine decarboxylase, structural 1
|
|
chr11_-_106993235
|
9.620
|
NM_176850
|
Bptf
|
bromodomain PHD finger transcription factor
|
|
chr4_-_120887671
|
9.524
|
|
Rlf
|
rearranged L-myc fusion sequence
|
|
chr18_+_75527015
|
9.482
|
NM_001042660
|
Smad7
|
MAD homolog 7 (Drosophila)
|
|
chrX_+_131135770
|
9.430
|
NM_019868
|
Hnrnph2
|
heterogeneous nuclear ribonucleoprotein H2
|
|
chr9_+_35075447
|
9.414
|
NM_028040
|
Rpusd4
|
RNA pseudouridylate synthase domain containing 4
|
|
chr15_+_34425046
|
9.348
|
NM_026340
NM_152894
|
Pop1
|
processing of precursor 1, ribonuclease P/MRP family, (S. cerevisiae)
|
|
chr18_+_12327237
|
9.225
|
NM_029623
|
3110002H16Rik
|
RIKEN cDNA 3110002H16 gene
|
|
chr9_+_106379599
|
9.219
|
NM_145620
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast)
|
|
chr9_-_22193627
|
9.208
|
NM_028390
|
Anln
|
anillin, actin binding protein
|
|
chrX_-_147531030
|
9.196
|
NM_001164578
NM_175146
|
Tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr10_-_77927652
|
9.144
|
NM_172134
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase
|
|
chr5_-_72559220
|
9.094
|
NM_178599
|
Commd8
|
COMM domain containing 8
|
|
chr11_-_69734131
|
9.072
|
NM_001166590
|
Eif5a
|
eukaryotic translation initiation factor 5A
|
|
chr3_-_19935200
|
9.054
|
|
Hps3
|
Hermansky-Pudlak syndrome 3 homolog (human)
|
|
chr5_+_28492235
|
9.042
|
NM_010134
|
En2
|
engrailed 2
|
|
chr18_-_80348164
|
9.035
|
NM_175028
|
Adnp2
|
ADNP homeobox 2
|
|
chr1_+_168309525
|
9.009
|
|
Tada1
|
transcriptional adaptor 1
|
|
chr1_-_130488785
|
8.938
|
NM_009911
|
Cxcr4
|
chemokine (C-X-C motif) receptor 4
|
|
chr8_-_86222963
|
8.933
|
NM_177262
|
Pkn1
|
protein kinase N1
|
|
chr11_-_93829704
|
8.877
|
NM_008704
|
Nme1
|
non-metastatic cells 1, protein (NM23A) expressed in
|
|
chr12_+_17551678
|
8.874
|
NM_013614
|
Odc1
|
ornithine decarboxylase, structural 1
|
|
chr1_+_182781247
|
8.834
|
NM_145943
|
BC031781
|
cDNA sequence BC031781
|
|
chr3_+_137527448
|
8.803
|
NM_016750
|
H2afz
|
H2A histone family, member Z
|
|
chr6_-_128312829
|
8.761
|
|
5930416I19Rik
|
RIKEN cDNA 5930416I19 gene
|
|
chr10_-_17742931
|
8.753
|
NM_028440
|
3110003A17Rik
|
RIKEN cDNA 3110003A17 gene
|
|
chr14_-_52816830
|
8.752
|
NM_033618
|
Supt16h
|
suppressor of Ty 16 homolog (S. cerevisiae)
|
|
chr17_-_33783578
|
8.735
|
|
Hnrnpm
|
heterogeneous nuclear ribonucleoprotein M
|
|
chr2_+_31325763
|
8.710
|
NM_007494
|
Ass1
|
argininosuccinate synthetase 1
|
|
chr5_+_147760198
|
8.675
|
NM_025652
|
Gtf3a
|
general transcription factor III A
|
|
chr7_-_51787440
|
8.666
|
NM_019866
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr8_+_96738490
|
8.633
|
NM_172410
|
Nup93
|
nucleoporin 93
|
|
chr16_+_44139908
|
8.630
|
NM_028108
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr17_-_24300592
|
8.613
|
NM_172935
|
Amdhd2
|
amidohydrolase domain containing 2
|
|
chrX_+_12858142
|
8.591
|
NM_010028
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr3_+_89518943
|
8.578
|
NM_001038587
|
Adar
|
adenosine deaminase, RNA-specific
|
|
chr10_-_62801726
|
8.562
|
NM_001159590
NM_019812
NM_001159589
|
Sirt1
|
sirtuin 1 (silent mating type information regulation 2, homolog) 1 (S. cerevisiae)
|
|
chr11_+_95198329
|
8.549
|
NM_172543
|
Fam117a
|
family with sequence similarity 117, memberA
|
|
chr19_-_40662819
|
8.509
|
NM_019698
NM_153554
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr5_+_31357119
|
8.483
|
NM_023525
|
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase
|
|
chr11_-_93829556
|
8.449
|
|
Nme1
|
non-metastatic cells 1, protein (NM23A) expressed in
|
|
chr11_-_17111513
|
8.411
|
|
Pno1
|
partner of NOB1 homolog (S. cerevisiae)
|
|
chr17_-_85189859
|
8.401
|
|
Lrpprc
|
leucine-rich PPR-motif containing
|
|
chr3_-_19935292
|
8.400
|
NM_001146323
NM_001146324
NM_080634
|
Hps3
|
Hermansky-Pudlak syndrome 3 homolog (human)
|
|
chr13_-_38750866
|
8.367
|
NM_025380
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1
|
|
chr3_-_129534032
|
8.360
|
|
Gar1
|
GAR1 ribonucleoprotein homolog (yeast)
|
|
chr9_-_95412264
|
8.343
|
NM_001114977
NM_026476
|
2610101N10Rik
|
RIKEN cDNA 2610101N10 gene
|
|
chr2_-_154394864
|
8.310
|
|
E2f1
|
E2F transcription factor 1
|
|
chr8_+_96738448
|
8.308
|
|
Nup93
|
nucleoporin 93
|
|
chrX_-_138159759
|
8.302
|
NM_010572
|
Irs4
|
insulin receptor substrate 4
|
|
chr8_-_126545208
|
8.260
|
NM_133966
|
Taf5l
|
TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor
|
|
chr12_-_80108263
|
8.238
|
NM_178745
|
Tmem229b
|
transmembrane protein 229B
|
|
chr5_-_100407613
|
8.234
|
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D
|
|
chr5_+_38611707
|
8.217
|
NM_025281
|
Lyar
|
Ly1 antibody reactive clone
|
|
chr15_-_98711692
|
8.157
|
NM_026967
|
Rhebl1
|
Ras homolog enriched in brain like 1
|
|
chr12_-_77470450
|
8.100
|
NM_001172104
NM_028356
|
Zbtb25
|
zinc finger and BTB domain containing 25
|