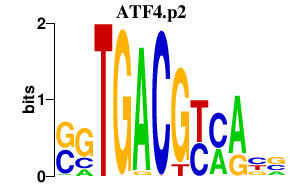
|
chr3_+_134875526
|
38.042
|
NM_173762
|
Cenpe
|
centromere protein E
|
|
chr5_-_130478834
|
36.510
|
|
Gusb
|
glucuronidase, beta
|
|
chr14_-_47896531
|
35.183
|
NM_172598
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr3_-_19211000
|
30.853
|
NM_001122759
|
Pde7a
|
phosphodiesterase 7A
|
|
chr17_+_35118956
|
30.614
|
NM_030597
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr3_-_36470904
|
30.395
|
NM_009828
|
Ccna2
|
cyclin A2
|
|
chr3_-_36470845
|
29.431
|
|
Ccna2
|
cyclin A2
|
|
chr1_-_71699671
|
29.027
|
NM_010233
|
Fn1
|
fibronectin 1
|
|
chr12_+_33505307
|
28.942
|
|
Nampt
|
nicotinamide phosphoribosyltransferase
|
|
chr14_-_47896464
|
28.866
|
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr3_-_36470882
|
28.386
|
|
Ccna2
|
cyclin A2
|
|
chr1_-_13650563
|
27.456
|
NM_145381
|
Lactb2
|
lactamase, beta 2
|
|
chr2_+_167514414
|
26.794
|
NM_009883
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr12_+_33505364
|
25.614
|
|
Nampt
|
nicotinamide phosphoribosyltransferase
|
|
chr18_+_11815869
|
25.323
|
|
Rbbp8
|
retinoblastoma binding protein 8
|
|
chr9_-_20781206
|
24.695
|
NM_010333
|
S1pr2
|
sphingosine-1-phosphate receptor 2
|
|
chr2_+_167514489
|
24.062
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr7_+_52973440
|
23.455
|
NM_009077
|
Rpl18
|
ribosomal protein L18
|
|
chr9_-_20781133
|
23.221
|
|
S1pr2
|
sphingosine-1-phosphate receptor 2
|
|
chr2_+_167514460
|
23.078
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr18_+_11816327
|
22.757
|
NM_001081223
|
Rbbp8
|
retinoblastoma binding protein 8
|
|
chr4_-_133801438
|
21.524
|
NM_024215
|
Zfp593
|
zinc finger protein 593
|
|
chr10_-_111434265
|
21.297
|
NM_028608
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr2_-_126959605
|
20.985
|
NM_144818
|
Ncaph
|
non-SMC condensin I complex, subunit H
|
|
chr15_+_79177959
|
20.896
|
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr4_-_55545211
|
20.852
|
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr8_+_77633426
|
20.741
|
NM_008566
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae)
|
|
chr10_+_85334563
|
20.370
|
NM_133993
|
Pwp1
|
PWP1 homolog (S. cerevisiae)
|
|
chr19_-_4625574
|
20.298
|
NM_023131
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr4_-_55545337
|
20.124
|
NM_010637
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr16_+_57549340
|
19.972
|
NM_001177871
|
Filip1l
|
filamin A interacting protein 1-like
|
|
chr17_-_74694322
|
19.786
|
|
Memo1
|
mediator of cell motility 1
|
|
chr3_-_107499129
|
18.994
|
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1
|
|
chr3_-_127256215
|
18.260
|
NM_138593
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr5_-_130478687
|
18.064
|
NM_010368
|
Gusb
|
glucuronidase, beta
|
|
chr9_-_59334393
|
18.058
|
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila)
|
|
chr14_-_67691221
|
17.940
|
NM_028032
|
Ppp2r2a
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), alpha isoform
|
|
chr15_+_61869529
|
17.906
|
|
Pvt1
|
plasmacytoma variant translocation 1
|
|
chr8_+_125000042
|
17.813
|
NM_153775
|
Ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe)
|
|
chr5_-_111082387
|
17.722
|
NM_153570
|
Noc4l
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr10_-_111434231
|
17.620
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr18_+_66618186
|
17.430
|
NM_021451
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr10_-_92185071
|
17.292
|
NM_008682
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated gene 1
|
|
chr17_-_71876150
|
17.281
|
NM_023294
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr17_-_71876127
|
17.133
|
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr15_+_79178063
|
16.943
|
NM_010755
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr8_+_74659107
|
16.899
|
NM_001001491
|
Tpm4
|
tropomyosin 4
|
|
chr12_-_103213890
|
16.823
|
|
Gm2701
|
predicted gene 2701
|
|
chr6_-_39675456
|
16.449
|
NM_139294
|
Braf
|
Braf transforming gene
|
|
chr8_-_87186691
|
16.061
|
NM_010499
|
Ier2
|
immediate early response 2
|
|
chr10_-_117283020
|
16.037
|
NM_024457
|
Rap1b
|
RAS related protein 1b
|
|
chr3_+_40603872
|
15.993
|
NM_011495
|
Plk4
|
polo-like kinase 4 (Drosophila)
|
|
chr17_+_75404865
|
15.934
|
NM_019919
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr5_-_111082269
|
15.730
|
|
Noc4l
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr11_+_83116148
|
15.526
|
NM_001035854
NM_027915
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr6_-_131243197
|
15.458
|
NM_025564
|
Magohb
|
mago-nashi homolog B (Drosophila)
|
|
chr2_-_132078887
|
15.453
|
NM_011045
|
Pcna
|
proliferating cell nuclear antigen
|
|
chr12_+_117519658
|
15.400
|
NM_028731
|
Esyt2
|
extended synaptotagmin-like protein 2
|
|
chrX_-_53851108
|
15.250
|
|
Mmgt1
|
membrane magnesium transporter 1
|
|
chr11_+_117710532
|
15.238
|
NM_001012273
NM_009689
|
Birc5
|
baculoviral IAP repeat-containing 5
|
|
chr19_-_34549624
|
15.071
|
NM_009890
|
Ch25h
|
cholesterol 25-hydroxylase
|
|
chr19_+_21346734
|
15.034
|
NM_009551
|
Zfand5
|
zinc finger, AN1-type domain 5
|
|
chr17_+_35118792
|
14.944
|
NM_001110101
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr7_-_31449991
|
14.445
|
NM_027999
|
Haus5
|
HAUS augmin-like complex, subunit 5
|
|
chr11_+_83116262
|
14.338
|
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr1_-_193007202
|
14.272
|
NM_007498
|
Atf3
|
activating transcription factor 3
|
|
chr17_-_43013320
|
14.225
|
NM_009847
|
Cd2ap
|
CD2-associated protein
|
|
chr17_-_53828596
|
14.185
|
NM_028232
|
Sgol1
|
shugoshin-like 1 (S. pombe)
|
|
chr5_-_152453753
|
14.014
|
NM_027009
|
Rfc3
|
replication factor C (activator 1) 3
|
|
chr5_+_112705715
|
14.011
|
NM_009419
|
Tpst2
|
protein-tyrosine sulfotransferase 2
|
|
chr11_-_77978936
|
14.010
|
NM_009423
|
Traf4
|
TNF receptor associated factor 4
|
|
chr3_+_137806411
|
13.959
|
NM_175389
|
Rg9mtd2
|
RNA (guanine-9-) methyltransferase domain containing 2
|
|
chr1_-_51535232
|
13.829
|
NM_028696
|
Obfc2a
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr11_+_76057230
|
13.767
|
NM_183263
|
Rnmtl1
|
RNA methyltransferase like 1
|
|
chr13_-_22134795
|
13.665
|
NM_178186
|
Hist1h2ag
Hist1h2ai
|
histone cluster 1, H2ag
histone cluster 1, H2ai
|
|
chr9_-_75257067
|
13.508
|
NM_027418
|
Mapk6
|
mitogen-activated protein kinase 6
|
|
chr10_-_117282901
|
13.480
|
|
Rap1b
|
RAS related protein 1b
|
|
chr16_+_24393435
|
13.343
|
NM_178665
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr8_+_131209614
|
13.242
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr17_-_34324234
|
13.169
|
NM_013585
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2)
|
|
chr17_-_35089399
|
13.053
|
|
1110038B12Rik
|
RIKEN cDNA 1110038B12 gene
|
|
chr14_-_69903072
|
12.995
|
NM_026331
|
Slc25a37
|
solute carrier family 25, member 37
|
|
chr7_+_25689359
|
12.935
|
NM_001130152
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr1_-_193006986
|
12.924
|
|
Atf3
|
activating transcription factor 3
|
|
chr2_-_132078946
|
12.757
|
|
Pcna
|
proliferating cell nuclear antigen
|
|
chrX_-_53851046
|
12.744
|
NM_146234
|
Mmgt1
|
membrane magnesium transporter 1
|
|
chr7_-_26035683
|
12.727
|
NM_010155
|
Erf
|
Ets2 repressor factor
|
|
chr7_-_31449951
|
12.659
|
|
Haus5
|
HAUS augmin-like complex, subunit 5
|
|
chr11_-_76057153
|
12.618
|
|
Glod4
|
glyoxalase domain containing 4
|
|
chr2_+_109120894
|
12.536
|
NM_139303
|
Kif18a
|
kinesin family member 18A
|
|
chr5_-_136410216
|
12.530
|
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr7_-_52781502
|
12.526
|
NM_008654
|
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr4_+_140857147
|
12.524
|
NM_010139
|
Epha2
|
Eph receptor A2
|
|
chr16_-_4077714
|
12.490
|
NM_026508
|
Trap1
|
TNF receptor-associated protein 1
|
|
chr19_+_6085096
|
12.479
|
NM_026410
|
Cdca5
|
cell division cycle associated 5
|
|
chr9_-_4309324
|
12.369
|
|
Aasdhppt
|
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase
|
|
chr6_+_146591082
|
12.313
|
NM_025315
|
Med21
|
mediator complex subunit 21
|
|
chr2_-_119980243
|
12.270
|
NM_133838
|
Ehd4
|
EH-domain containing 4
|
|
chr19_+_7568529
|
12.221
|
NM_001163505
|
Atl3
|
atlastin GTPase 3
|
|
chr9_+_108462759
|
12.152
|
NM_011830
|
Impdh2
|
inosine 5'-phosphate dehydrogenase 2
|
|
chr8_-_82235601
|
12.028
|
NM_015751
|
Abce1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chr2_-_119980210
|
11.986
|
|
Ehd4
|
EH-domain containing 4
|
|
chr17_+_75404893
|
11.958
|
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chrX_+_71558358
|
11.913
|
NM_138607
|
Fam50a
|
family with sequence similarity 50, member A
|
|
chr5_+_35104416
|
11.888
|
|
Htt
|
huntingtin
|
|
chr3_-_127256181
|
11.720
|
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr2_+_164571458
|
11.660
|
NM_133763
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr17_-_74694120
|
11.556
|
NM_133771
|
Memo1
|
mediator of cell motility 1
|
|
chr7_+_25689409
|
11.465
|
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1
|
|
chr18_-_52688607
|
11.448
|
|
Lox
|
lysyl oxidase
|
|
chr10_-_17667872
|
11.413
|
NM_001033432
|
Heca
|
headcase homolog (Drosophila)
|
|
chr9_+_108957402
|
11.401
|
NM_026381
|
Shisa5
|
shisa homolog 5 (Xenopus laevis)
|
|
chr17_-_26206487
|
11.255
|
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II)
|
|
chr5_+_123526279
|
11.125
|
NM_001039723
|
Tmem120b
|
transmembrane protein 120B
|
|
chr8_+_119445639
|
11.077
|
NM_028131
|
Cenpn
|
centromere protein N
|
|
chr19_+_6057884
|
11.043
|
NM_001160239
NM_007990
NM_001190436
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived)
|
|
chr8_+_131209680
|
11.026
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr7_+_28954717
|
11.026
|
NM_007991
|
Fbl
|
fibrillarin
|
|
chr2_+_156665812
|
11.004
|
NM_173396
|
Tgif2
|
TGFB-induced factor homeobox 2
|
|
chr2_-_179777080
|
10.915
|
NM_011969
|
Psma7
|
proteasome (prosome, macropain) subunit, alpha type 7
|
|
chr8_-_108535317
|
10.875
|
NM_028038
|
Ddx28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr3_+_95462640
|
10.713
|
NM_008562
|
Mcl1
|
myeloid cell leukemia sequence 1
|
|
chr3_+_95462705
|
10.577
|
|
Mcl1
|
myeloid cell leukemia sequence 1
|
|
chr6_+_87863915
|
10.522
|
NM_173737
|
8430410A17Rik
|
RIKEN cDNA 8430410A17 gene
|
|
chr9_-_44152208
|
10.402
|
|
Hmbs
|
hydroxymethylbilane synthase
|
|
chr11_-_69485842
|
10.388
|
NM_001159375
NM_144958
|
Eif4a1
|
eukaryotic translation initiation factor 4A1
|
|
chr3_-_143868210
|
10.372
|
NM_010723
|
Lmo4
|
LIM domain only 4
|
|
chr16_-_33967064
|
10.358
|
NM_009471
|
Umps
|
uridine monophosphate synthetase
|
|
chr7_-_20214596
|
10.333
|
NM_009046
|
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B
|
|
chr7_-_72017803
|
10.331
|
NM_023239
|
Ndnl2
|
necdin-like 2
|
|
chr2_+_118872854
|
10.294
|
NM_029617
|
Casc5
|
cancer susceptibility candidate 5
|
|
chr3_+_30993952
|
10.284
|
NM_001039090
NM_011386
|
Skil
|
SKI-like
|
|
chr6_+_140572056
|
10.241
|
NM_001005605
|
Aebp2
|
AE binding protein 2
|
|
chr8_+_131209787
|
10.153
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr2_+_32631305
|
10.103
|
NM_178595
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr5_-_4104331
|
10.068
|
|
Cyp51
|
cytochrome P450, family 51
|
|
chr17_+_15841889
|
10.042
|
NM_007690
|
Chd1
|
chromodomain helicase DNA binding protein 1
|
|
chrX_-_35917684
|
10.026
|
|
Cul4b
|
cullin 4B
|
|
chr3_-_143868114
|
10.026
|
|
Lmo4
|
LIM domain only 4
|
|
chr7_-_126864266
|
9.990
|
NM_145585
|
Thumpd1
|
THUMP domain containing 1
|
|
chr4_+_129784799
|
9.985
|
NM_026441
|
Pef1
|
penta-EF hand domain containing 1
|
|
chr15_+_100058249
|
9.945
|
NM_007497
|
Atf1
|
activating transcription factor 1
|
|
chr9_-_119231439
|
9.906
|
|
Oxsr1
|
oxidative-stress responsive 1
|
|
chr11_-_69485800
|
9.882
|
|
Eif4a1
|
eukaryotic translation initiation factor 4A1
|
|
chr11_-_70051081
|
9.874
|
|
|
|
|
chr1_+_137625754
|
9.871
|
NM_007791
|
Csrp1
|
cysteine and glycine-rich protein 1
|
|
chr5_+_33361386
|
9.829
|
NM_011738
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide
|
|
chr19_-_61304260
|
9.817
|
NM_009970
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr11_-_76057188
|
9.796
|
NM_026029
|
Glod4
|
glyoxalase domain containing 4
|
|
chr17_-_57150821
|
9.779
|
|
Gtf2f1
|
general transcription factor IIF, polypeptide 1
|
|
chr15_+_65618577
|
9.765
|
NM_133766
|
Efr3a
|
EFR3 homolog A (S. cerevisiae)
|
|
chr17_+_34324636
|
9.735
|
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr5_+_111082466
|
9.728
|
NM_027156
|
Ddx51
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51
|
|
chr7_+_3655919
|
9.685
|
|
Rps9
|
ribosomal protein S9
|
|
chr14_-_47809063
|
9.516
|
NM_008102
|
Gch1
|
GTP cyclohydrolase 1
|
|
chr3_-_108643390
|
9.482
|
NM_011504
|
Stxbp3a
Stxbp3b
|
syntaxin binding protein 3A
syntaxin-binding protein 3B
|
|
chr16_-_57606866
|
9.438
|
NM_025599
|
2610528E23Rik
|
RIKEN cDNA 2610528E23 gene
|
|
chr8_-_4779533
|
9.426
|
NM_011369
|
Shcbp1
|
Shc SH2-domain binding protein 1
|
|
chr8_-_41596638
|
9.398
|
NM_011135
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7
|
|
chrX_+_151696991
|
9.395
|
NM_019736
|
Acot9
|
acyl-CoA thioesterase 9
|
|
chr5_+_3343861
|
9.375
|
|
Cdk6
|
cyclin-dependent kinase 6
|
|
chr10_+_96079634
|
9.363
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr11_-_106117095
|
9.350
|
NM_025310
|
Ftsj3
|
FtsJ homolog 3 (E. coli)
|
|
chr17_+_23816331
|
9.158
|
NM_018777
|
Cldn6
|
claudin 6
|
|
chr19_+_37450854
|
9.157
|
NM_010615
|
Kif11
|
kinesin family member 11
|
|
chr7_+_54101157
|
9.104
|
NM_010699
|
Ldha
|
lactate dehydrogenase A
|
|
chr9_-_44152242
|
9.093
|
NM_013551
|
Hmbs
|
hydroxymethylbilane synthase
|
|
chr12_+_70473032
|
9.062
|
NM_007481
|
Arf6
|
ADP-ribosylation factor 6
|
|
chr5_+_143722032
|
9.010
|
NM_007984
|
Fscn1
|
fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus)
|
|
chr4_+_126354857
|
8.969
|
NM_011970
|
Psmb2
|
proteasome (prosome, macropain) subunit, beta type 2
|
|
chr14_+_47896669
|
8.923
|
NM_080843
|
Socs4
|
suppressor of cytokine signaling 4
|
|
chr5_-_4104696
|
8.909
|
NM_020010
|
Cyp51
|
cytochrome P450, family 51
|
|
chr2_-_23011403
|
8.856
|
NM_025979
|
Mastl
|
microtubule associated serine/threonine kinase-like
|
|
chr5_+_151325197
|
8.818
|
NM_009765
NM_001081001
|
Brca2
|
breast cancer 2
|
|
chr15_-_20596495
|
8.777
|
NM_022816
|
Acot10
|
acyl-CoA thioesterase 10
|
|
chr19_-_6057706
|
8.761
|
NM_026246
|
Mrpl49
|
mitochondrial ribosomal protein L49
|
|
chr19_+_37000568
|
8.750
|
NM_001080706
|
Btaf1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, (Mot1 homolog, S. cerevisiae)
|
|
chr3_-_127256114
|
8.749
|
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr17_+_34324700
|
8.709
|
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr10_-_75322941
|
8.639
|
NM_010798
|
Mif
|
macrophage migration inhibitory factor
|
|
chr17_-_26206459
|
8.592
|
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II)
|
|
chr13_+_111185251
|
8.442
|
NM_152804
|
Plk2
|
polo-like kinase 2 (Drosophila)
|
|
chr6_-_148893155
|
8.413
|
|
Fam60a
|
family with sequence similarity 60, member A
|
|
chr9_+_113650560
|
8.391
|
NM_001114347
|
Clasp2
|
CLIP associating protein 2
|
|
chr1_+_95375503
|
8.360
|
NM_001159717
NM_001159718
NM_010891
NM_001159719
|
Sept2
|
septin 2
|
|
chr1_-_193399412
|
8.326
|
NM_029766
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr5_+_125029125
|
8.309
|
NM_181410
|
Gtf2h3
|
general transcription factor IIH, polypeptide 3
|
|
chr2_+_146838735
|
8.292
|
NM_011917
|
Xrn2
|
5'-3' exoribonuclease 2
|
|
chr7_+_3655602
|
8.292
|
NM_029767
|
Rps9
|
ribosomal protein S9
|
|
chr8_-_72149070
|
8.288
|
NM_001045553
|
Zfp868
|
zinc finger protein 868
|
|
chr6_+_120786323
|
8.281
|
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr10_+_82092582
|
8.247
|
NM_011561
NM_172552
|
Tdg
Gm9855
|
thymine DNA glycosylase
thymine DNA glycosylase pseudogene
|
|
chr9_-_97011440
|
8.240
|
NM_138756
|
Slc25a36
|
solute carrier family 25, member 36
|
|
chr8_-_72498125
|
8.205
|
|
Gatad2a
|
GATA zinc finger domain containing 2A
|
|
chr4_-_136913576
|
8.204
|
NM_009861
|
Cdc42
|
cell division cycle 42 homolog (S. cerevisiae)
|
|
chr19_-_4625276
|
8.164
|
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr1_-_167167019
|
8.160
|
NM_145513
|
Tiprl
|
TIP41, TOR signalling pathway regulator-like (S. cerevisiae)
|
|
chr1_-_95698465
|
8.156
|
NM_001105667
NM_023136
|
Dtymk
|
deoxythymidylate kinase
|
|
chr10_-_119638448
|
8.127
|
NM_028679
|
Irak3
|
interleukin-1 receptor-associated kinase 3
|
|
chr12_-_74387687
|
8.106
|
NM_029580
|
Trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae)
|