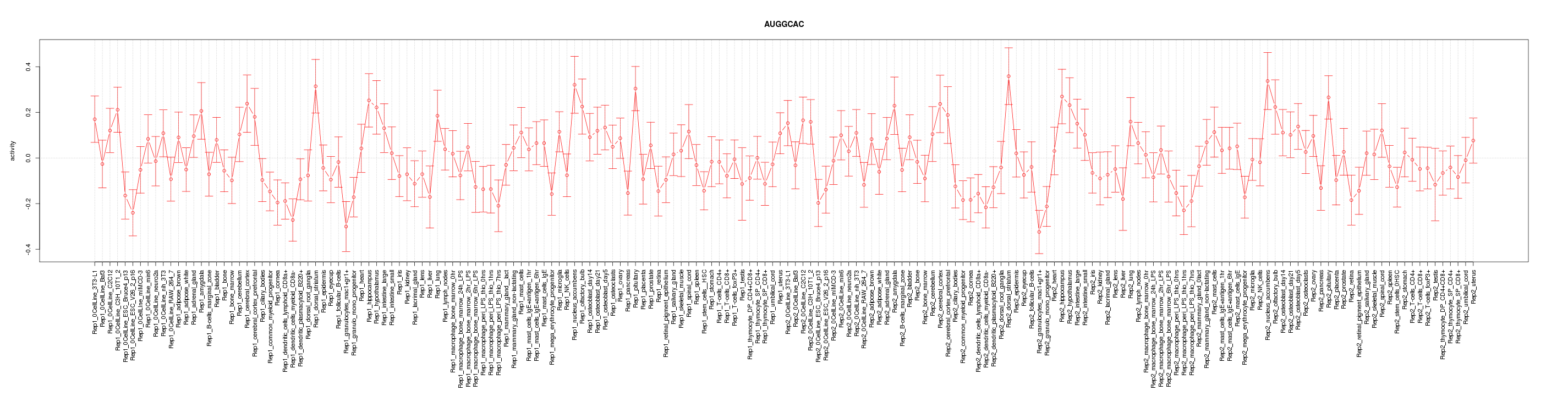
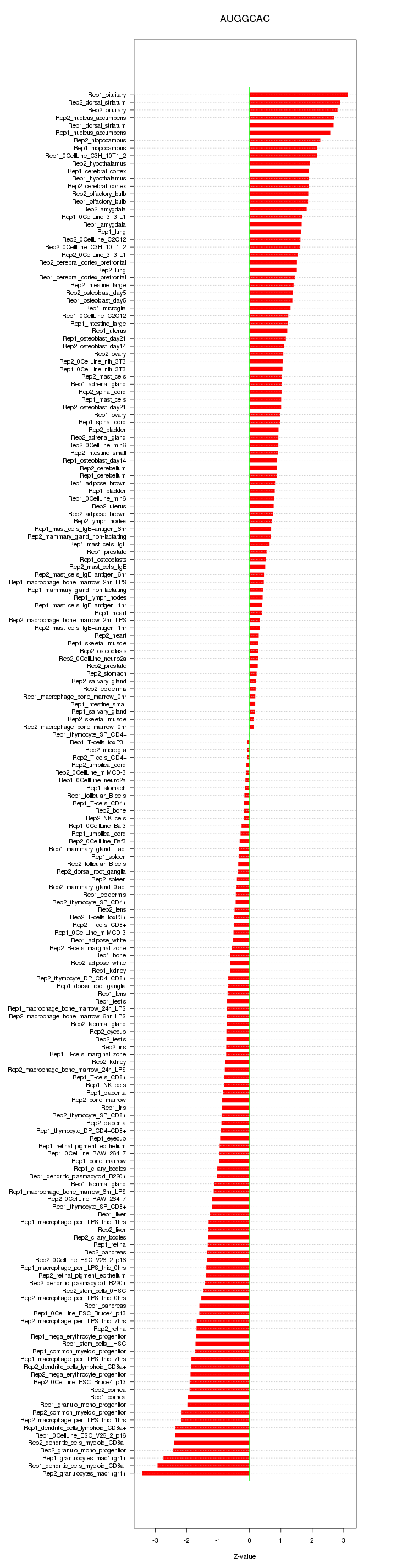
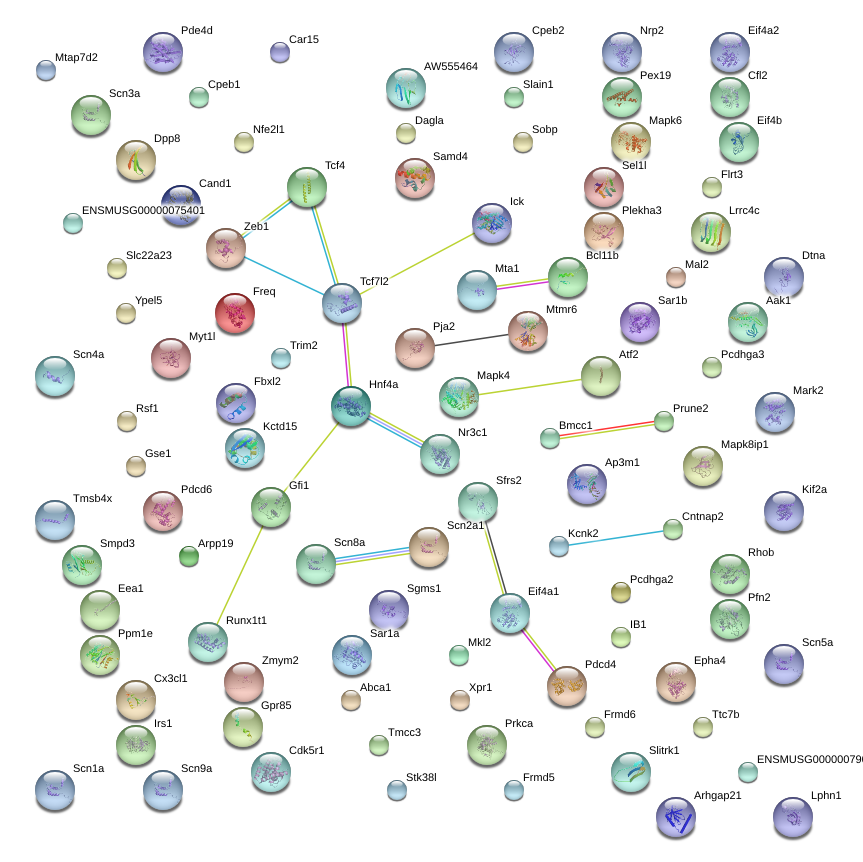
|
chr3_-_57651621
|
34.534
|
NM_019410
|
Pfn2
|
profilin 2
|
|
chr1_-_191167231
|
27.938
|
NM_001159850
|
Kcnk2
|
potassium channel, subfamily K, member 2
|
|
chr1_-_191167582
|
26.470
|
NM_010607
|
Kcnk2
|
potassium channel, subfamily K, member 2
|
|
chr8_+_97296066
|
22.455
|
NM_009142
|
Cx3cl1
|
chemokine (C-X3-C motif) ligand 1
|
|
chr18_+_37833939
|
21.554
|
NM_033586
|
Pcdhga3
|
protocadherin gamma subfamily A, 3
|
|
chr18_+_37844927
|
21.348
|
NM_033587
|
Pcdhga4
|
protocadherin gamma subfamily A, 4
|
|
chr18_+_37828606
|
21.181
|
NM_033585
|
Pcdhga2
|
protocadherin gamma subfamily A, 2
|
|
chr18_+_37885359
|
21.033
|
NM_033591
|
Pcdhga8
|
protocadherin gamma subfamily A, 8
|
|
chr18_+_37906739
|
20.747
|
NM_033593
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr18_+_37890662
|
20.542
|
NM_033577
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5
|
|
chr18_+_37966017
|
20.368
|
NM_033581
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3
|
|
chr18_+_37915375
|
20.262
|
NM_033594
|
Pcdhga11
|
protocadherin gamma subfamily A, 11
|
|
chr18_+_37839910
|
20.046
|
NM_033574
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1
|
|
chr18_+_37925233
|
19.800
|
NM_033595
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr18_+_37901716
|
19.734
|
NM_033578
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6
|
|
chr18_+_37874471
|
19.729
|
NM_033590
|
Pcdhga7
|
protocadherin gamma subfamily A, 7
|
|
chr18_+_37896589
|
19.698
|
NM_033592
|
Pcdhga9
|
protocadherin gamma subfamily A, 9
|
|
chr18_+_37911236
|
19.498
|
NM_033579
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7
|
|
chr18_+_37921454
|
19.323
|
NM_033580
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8
|
|
chr18_+_37821598
|
18.985
|
NM_033584
|
Pcdhga1
|
protocadherin gamma subfamily A, 1
|
|
chr18_+_37866747
|
18.606
|
NM_033589
|
Pcdhga6
|
protocadherin gamma subfamily A, 6
|
|
chr18_+_37880036
|
18.469
|
NM_033576
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4
|
|
chr15_+_54402914
|
17.900
|
NM_178920
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr18_+_37979199
|
17.651
|
NM_033583
|
Pcdhgb4
Pcdhgc5
|
protocadherin gamma subfamily B, 4
protocadherin gamma subfamily C, 5
|
|
chr18_+_37849488
|
17.451
|
NM_033575
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2
|
|
chr18_+_37974732
|
17.348
|
NM_033582
|
Pcdhgc4
|
protocadherin gamma subfamily C, 4
|
|
chr18_+_37854011
|
17.150
|
NM_033588
|
Pcdhga5
|
protocadherin gamma subfamily A, 5
|
|
chr13_+_109444370
|
14.998
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr2_-_92241357
|
14.819
|
NM_011162
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr11_-_108205199
|
13.779
|
NM_011101
|
Prkca
|
protein kinase C, alpha
|
|
chr3_-_84024766
|
13.499
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr11_-_87172419
|
12.077
|
NM_177167
|
Ppm1e
|
protein phosphatase 1E (PP2C domain containing)
|
|
chr9_-_113935832
|
11.581
|
NM_178624
|
Fbxl2
|
F-box and leucine-rich repeat protein 2
|
|
chr2_-_20889276
|
10.870
|
NM_001081364
NM_001128084
|
Arhgap21
|
Rho GTPase activating protein 21
|
|
chr12_-_93087514
|
10.705
|
NM_001039089
NM_011344
|
Sel1l
|
sel-1 suppressor of lin-12-like (C. elegans)
|
|
chr13_-_34437042
|
10.283
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr14_-_109313447
|
8.799
|
NM_199065
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr14_+_47621014
|
8.789
|
NM_001163433
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr8_+_86423968
|
8.668
|
NM_181039
|
Lphn1
|
latrophilin 1
|
|
chrX_+_155852439
|
8.506
|
NM_001081124
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr18_+_23573908
|
8.321
|
NM_010087
NM_207650
|
Dtna
|
dystrobrevin alpha
|
|
chr7_-_88599560
|
7.853
|
NM_007755
|
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1
|
|
chr18_-_74224541
|
7.162
|
NM_172632
|
Mapk4
|
mitogen-activated protein kinase 4
|
|
chr1_+_174056868
|
7.157
|
NM_001159525
NM_023041
|
Pex19
|
peroxisomal biogenesis factor 19
|
|
chr9_+_74885522
|
6.976
|
NM_021548
|
Arpp19
|
cAMP-regulated phosphoprotein 19
|
|
chr9_+_74885420
|
6.862
|
NM_001142655
|
Arpp19
|
cAMP-regulated phosphoprotein 19
|
|
chr2_+_97307652
|
6.797
|
NM_178725
|
Lrrc4c
|
leucine rich repeat containing 4C
|
|
chr10_+_94039007
|
6.766
|
NM_177026
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr2_+_76513326
|
6.738
|
NM_031256
|
Plekha3
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 3
|
|
chr10_+_94038001
|
6.640
|
NM_001168684
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr17_-_64681166
|
6.491
|
NM_001025309
NM_144859
|
Pja2
|
praja 2, RING-H2 motif containing
|
|
chr12_+_71926429
|
6.431
|
NM_028127
|
Frmd6
|
FERM domain containing 6
|
|
chr12_-_101758964
|
6.420
|
NM_001033213
|
Ttc7b
|
tetratricopeptide repeat domain 7B
|
|
chr19_-_10379361
|
6.057
|
NM_198114
|
Dagla
|
diacylglycerol lipase, alpha
|
|
chr12_-_8506790
|
6.046
|
NM_007483
|
Rhob
|
ras homolog gene family, member B
|
|
chr9_+_64880208
|
5.888
|
NM_028906
|
Dpp8
|
dipeptidylpeptidase 8
|
|
chr2_+_65508501
|
5.846
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chr2_-_73730605
|
5.737
|
NM_001025093
NM_009715
|
Atf2
|
activating transcription factor 2
|
|
chr18_+_69505374
|
5.710
|
NM_001083967
|
Tcf4
|
transcription factor 4
|
|
chr12_+_30219769
|
5.447
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr2_-_140497204
|
5.072
|
NM_001172160
NM_178382
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr10_+_95403283
|
5.008
|
NM_001001932
|
Eea1
|
early endosome antigen 1
|
|
chr16_+_23107538
|
4.998
|
NM_001123037
NM_001123038
NM_013506
|
Eif4a2
|
eukaryotic translation initiation factor 4A2
|
|
chr18_+_69504145
|
4.768
|
NM_013685
|
Tcf4
|
transcription factor 4
|
|
chr4_+_13711928
|
4.661
|
NM_009822
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chrX_-_163647088
|
4.652
|
NM_021278
|
Tmsb4x
|
thymosin, beta 4, X chromosome
|
|
chr12_+_114336416
|
4.445
|
NM_054081
|
Mta1
|
metastasis associated 1
|
|
chr8_-_108861850
|
4.357
|
NM_021491
|
Smpd3
|
sphingomyelin phosphodiesterase 3, neutral
|
|
chr6_+_47194393
|
4.220
|
NM_025771
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr14_+_47502637
|
4.188
|
NM_001037221
NM_028966
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr10_-_118677059
|
4.159
|
NM_027994
|
Cand1
|
cullin associated and neddylation disassociated 1
|
|
chr12_+_30213224
|
4.062
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr18_+_5591869
|
3.869
|
NM_011546
|
Zeb1
|
zinc finger E-box binding homeobox 1
|
|
chr11_-_116714406
|
3.692
|
NM_011358
|
Srsf2
|
serine/arginine-rich splicing factor 2
|
|
chr4_+_13670425
|
3.664
|
NM_001111026
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr14_-_21871647
|
3.626
|
NM_018829
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr10_+_93977597
|
3.569
|
NM_172051
|
Tmcc3
|
transmembrane and coiled coil domains 3
|
|
chr7_-_35437859
|
3.253
|
NM_146188
|
Kctd15
|
potassium channel tetramerisation domain containing 15
|
|
chr6_+_45010040
|
3.168
|
NM_001004357
|
Cntnap2
|
contactin associated protein-like 2
|
|
chr13_+_105607262
|
3.089
|
NM_026075
|
Srek1ip1
|
splicing regulatory glutamine/lysine-rich protein 1interacting protein 1
|
|
chr12_-_109241423
|
3.064
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr1_-_77511653
|
3.047
|
NM_007936
|
Epha4
|
Eph receptor A4
|
|
chr11_-_96691261
|
2.878
|
NM_008686
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1
|
|
chr11_-_96690784
|
2.831
|
NM_001130450
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1
|
|
chr19_+_53966720
|
2.704
|
NM_011050
|
Pdcd4
|
programmed cell death 4
|
|
chr18_-_39646734
|
2.589
|
NM_008173
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr2_+_31101302
|
2.568
|
NM_019681
|
Ncs1
|
neuronal calcium sensor 1
|
|
chr12_+_113960379
|
2.553
|
NM_001024602
|
AW555464
|
expressed sequence AW555464
|
|
chr16_-_17891608
|
2.550
|
NM_001109750
NM_010048
|
Dgcr2
|
DiGeorge syndrome critical region gene 2
|
|
chr12_-_55963776
|
2.528
|
NM_007688
|
Cfl2
|
cofilin 2, muscle
|
|
chr19_-_7369859
|
2.504
|
NM_001080390
|
Mark2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr19_+_17030600
|
2.437
|
NM_181348
|
Prune2
|
prune homolog 2 (Drosophila)
|
|
chr16_+_13358513
|
2.429
|
NM_153588
|
Mkl2
|
MKL/myocardin-like 2
|
|
chr14_+_60884038
|
2.410
|
NM_144843
|
Mtmr6
|
myotubularin related protein 6
|
|
chr1_-_82287990
|
2.307
|
NM_010570
|
Irs1
|
insulin receptor substrate 1
|
|
chr4_-_53172748
|
2.139
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_-_65405546
|
2.062
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr17_+_73185961
|
1.907
|
NM_027166
|
Ypel5
|
yippee-like 5 (Drosophila)
|
|
chr16_-_87432721
|
1.852
|
NM_001081068
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1
|
|
chr13_-_74454770
|
1.799
|
NM_011051
|
Pdcd6
|
programmed cell death 6
|
|
chr8_+_123012667
|
1.698
|
NM_198671
|
Gse1
|
genetic suppressor element 1
|
|
chr10_-_42894259
|
1.693
|
NM_175407
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr16_+_13256573
|
1.621
|
NM_001122667
|
Mkl2
|
MKL/myocardin-like 2
|
|
chr1_-_157264398
|
1.598
|
NM_011273
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1
|
|
chr14_+_104049453
|
1.592
|
NM_198014
|
Slain1
|
SLAIN motif family, member 1
|
|
chr6_+_86799460
|
1.576
|
NM_001040106
NM_177762
|
Aak1
|
AP2 associated kinase 1
|
|
chr2_-_121632749
|
1.512
|
NM_172673
|
Frmd5
|
FERM domain containing 5
|
|
chr11_+_80290509
|
1.511
|
NM_009871
|
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr6_+_146673451
|
1.462
|
NM_172734
|
Stk38l
|
serine/threonine kinase 38 like
|
|
chr6_-_13789847
|
1.384
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr10_+_61143056
|
1.298
|
NM_009120
|
Sar1a
|
SAR1 gene homolog A (S. cerevisiae)
|
|
chr9_+_77956998
|
1.272
|
NM_001163780
|
Ick
|
intestinal cell kinase
|
|
chr5_+_43624701
|
1.270
|
NM_001177379
NM_175937
|
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2
|
|
chr14_+_57506630
|
1.260
|
NM_029498
|
Zmym2
|
zinc finger, MYM-type 2
|
|
chr7_+_104728398
|
1.257
|
NM_001081267
|
Rsf1
|
remodeling and spacing factor 1
|
|
chr19_-_32285445
|
1.194
|
NM_001168526
|
Sgms1
|
sphingomyelin synthase 1
|
|
chr1_+_62749850
|
1.172
|
NM_001077403
NM_001077404
NM_001077405
NM_001077406
NM_001077407
NM_010939
|
Nrp2
|
neuropilin 2
|
|
chr13_-_107812106
|
1.100
|
NM_001145779
NM_008442
|
Kif2a
|
kinesin family member 2A
|
|
chr6_-_12699189
|
1.084
|
NM_001164805
|
Thsd7a
|
thrombospondin, type I, domain containing 7A
|
|
chr19_-_7416286
|
1.072
|
NM_001080388
NM_001080389
NM_007928
|
Mark2
|
MAP/microtubule affinity-regulating kinase 2
|
|
chr3_-_72860864
|
1.070
|
NM_198864
|
Slitrk3
|
SLIT and NTRK-like family, member 3
|
|
chr14_-_66043991
|
1.019
|
NM_010932
|
Pnoc
|
prepronociceptin
|
|
chr7_+_107855215
|
0.996
|
NM_178764
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr4_+_57580989
|
0.984
|
NM_172868
|
Palm2
|
paralemmin 2
|
|
chr7_-_118089700
|
0.964
|
NM_010826
|
Mrvi1
|
MRV integration site 1
|
|
chr13_-_18035913
|
0.959
|
NM_019491
|
Rala
|
v-ral simian leukemia viral oncogene homolog A (ras related)
|
|
chr17_-_80295336
|
0.943
|
NM_019717
NM_178050
|
Atl2
|
atlastin GTPase 2
|
|
chr2_+_58998885
|
0.891
|
NM_026361
NM_175464
|
Pkp4
|
plakophilin 4
|
|
chr3_-_40886957
|
0.869
|
NM_027558
|
Pgrmc2
|
progesterone receptor membrane component 2
|
|
chr5_+_9266137
|
0.820
|
NM_172706
|
9330182L06Rik
|
RIKEN cDNA 9330182L06 gene
|
|
chr12_-_99816091
|
0.782
|
NM_029911
|
Kcnk10
|
potassium channel, subfamily K, member 10
|
|
chr9_-_36954858
|
0.745
|
NM_148950
|
Pknox2
|
Pbx/knotted 1 homeobox 2
|
|
chr8_+_34710049
|
0.685
|
NM_017374
|
Ppp2cb
|
protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform
|
|
chr7_-_118125571
|
0.678
|
NM_194464
|
Mrvi1
|
MRV integration site 1
|
|
chr19_+_25311691
|
0.673
|
NM_181404
|
Kank1
|
KN motif and ankyrin repeat domains 1
|
|
chr19_+_53977788
|
0.659
|
NM_001168491
NM_001168492
|
Pdcd4
|
programmed cell death 4
|
|
chr4_-_34634507
|
0.657
|
NM_011895
|
Slc35a1
|
solute carrier family 35 (CMP-sialic acid transporter), member 1
|
|
chr7_+_116154373
|
0.572
|
NM_021885
|
Tub
|
tubby candidate gene
|
|
chr8_+_129587894
|
0.506
|
NM_001122850
NM_033620
|
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans)
|
|
chr11_+_51912319
|
0.355
|
NM_019411
|
Ppp2ca
|
protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform
|
|
chr7_+_16715637
|
0.354
|
NM_148946
|
Slc8a2
|
solute carrier family 8 (sodium/calcium exchanger), member 2
|
|
chr9_-_36954842
|
0.340
|
NM_001029838
|
Pknox2
|
Pbx/knotted 1 homeobox 2
|
|
chr13_+_37980739
|
0.300
|
NM_001177869
|
Rreb1
|
ras responsive element binding protein 1
|
|
chr9_-_86465373
|
0.280
|
NM_001163746
NM_028352
|
Pgm3
|
phosphoglucomutase 3
|
|
chr3_+_30894668
|
0.277
|
NM_008857
|
Prkci
|
protein kinase C, iota
|
|
chr6_-_134516930
|
0.265
|
NM_008514
|
Lrp6
|
low density lipoprotein receptor-related protein 6
|
|
chr8_+_123061308
|
0.255
|
NM_001145896
|
Gse1
|
genetic suppressor element 1
|
|
chr8_+_129823507
|
0.249
|
NM_001013581
NM_001013580
|
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans)
|
|
chr19_-_32423805
|
0.039
|
NM_001168525
|
Sgms1
|
sphingomyelin synthase 1
|