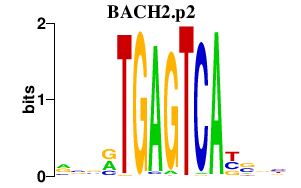
|
chr9_-_86589782
|
35.675
|
NM_008615
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr4_-_129851289
|
34.948
|
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr7_+_135667095
|
24.645
|
NM_013863
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr7_+_135667279
|
24.187
|
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr1_+_173367685
|
23.773
|
NM_172647
|
F11r
|
F11 receptor
|
|
chr11_-_55223449
|
23.589
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr7_+_31337558
|
23.315
|
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr12_+_31950252
|
22.272
|
|
Lamb1
|
laminin B1
|
|
chr7_-_112630662
|
22.049
|
NM_028444
|
Prkcdbp
|
protein kinase C, delta binding protein
|
|
chr12_+_31950097
|
21.794
|
NM_008482
|
Lamb1
|
laminin B1
|
|
chr9_+_44850426
|
21.298
|
NM_007962
|
Mpzl2
|
myelin protein zero-like 2
|
|
chr7_+_31338319
|
20.589
|
NM_001012401
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr7_-_116688776
|
20.553
|
|
St5
|
suppression of tumorigenicity 5
|
|
chr7_-_30033397
|
20.011
|
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene
|
|
chr19_-_38199291
|
20.011
|
NM_011255
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr7_+_138087002
|
19.927
|
|
Htra1
|
HtrA serine peptidase 1
|
|
chr19_-_38199036
|
18.754
|
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr18_+_61204798
|
18.456
|
NM_001146268
NM_008809
|
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide
|
|
chr3_-_88297220
|
18.331
|
NM_019390
|
Lmna
|
lamin A
|
|
chr11_+_78001871
|
18.058
|
NM_001159482
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr6_-_137468412
|
17.652
|
|
Eps8
|
epidermal growth factor receptor pathway substrate 8
|
|
chr7_+_148654681
|
17.257
|
NM_001111050
|
Cd151
|
CD151 antigen
|
|
chr2_+_155215225
|
16.897
|
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2
|
|
chr2_+_167514489
|
16.399
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr7_-_51926142
|
16.377
|
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr3_-_95543477
|
16.190
|
|
Ecm1
|
extracellular matrix protein 1
|
|
chr3_-_95543488
|
16.107
|
|
Ecm1
|
extracellular matrix protein 1
|
|
chr14_+_102239673
|
15.864
|
|
Lmo7
|
LIM domain only 7
|
|
chr3_+_93324411
|
15.734
|
NM_016740
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin)
|
|
chr3_-_95543490
|
15.374
|
NM_007899
|
Ecm1
|
extracellular matrix protein 1
|
|
chr19_-_38199267
|
15.004
|
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr3_-_92289795
|
14.906
|
NM_009264
|
Sprr1a
|
small proline-rich protein 1A
|
|
chr9_+_44850616
|
14.877
|
|
Mpzl2
|
myelin protein zero-like 2
|
|
chr12_-_104970691
|
14.723
|
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chr11_+_94797565
|
14.549
|
NM_007742
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr11_+_76718046
|
14.403
|
NM_025655
|
Tmigd1
|
transmembrane and immunoglobulin domain containing 1
|
|
chr10_+_59740373
|
14.360
|
NM_001146120
NM_001146121
NM_001146122
NM_001146123
NM_011179
|
Psap
|
prosaposin
|
|
chr2_+_143741050
|
14.333
|
NM_019771
|
Dstn
|
destrin
|
|
chr17_-_24781872
|
14.322
|
NM_023449
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2
|
|
chr2_+_35159780
|
14.159
|
|
Gsn
|
gelsolin
|
|
chr11_+_78002309
|
13.812
|
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr11_-_100259061
|
13.639
|
|
Jup
|
junction plakoglobin
|
|
chr10_+_59740430
|
13.602
|
|
Psap
|
prosaposin
|
|
chr6_+_17413930
|
13.601
|
NM_008591
|
Met
|
met proto-oncogene
|
|
chr10_-_76188683
|
13.502
|
NM_009933
|
Col6a1
|
collagen, type VI, alpha 1
|
|
chr4_+_118199847
|
13.335
|
NM_025572
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene
|
|
chr3_-_88307198
|
13.333
|
|
Lmna
|
lamin A
|
|
chr8_-_64069935
|
13.331
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr12_-_105101768
|
13.176
|
|
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A
|
|
chr10_+_18564876
|
13.050
|
NM_022032
|
Perp
|
PERP, TP53 apoptosis effector
|
|
chr3_+_93324437
|
12.631
|
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin)
|
|
chr14_+_66587319
|
12.539
|
NM_013492
|
Clu
|
clusterin
|
|
chrX_+_93650774
|
12.463
|
NM_001159628
NM_010417
|
Heph
|
hephaestin
|
|
chr9_+_108241754
|
12.408
|
|
Gpx1
|
glutathione peroxidase 1
|
|
chr16_+_24448176
|
12.177
|
NM_001145952
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr3_-_88307253
|
12.165
|
NM_001002011
NM_001111102
|
Lmna
|
lamin A
|
|
chr12_-_105195097
|
12.139
|
NM_009247
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E
|
|
chr10_-_127935725
|
11.934
|
NM_172259
|
Myl6b
|
myosin, light polypeptide 6B
|
|
chr15_-_83002624
|
11.763
|
NM_029787
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr9_+_69301471
|
11.733
|
NM_007585
|
Anxa2
|
annexin A2
|
|
chr6_-_113384474
|
11.638
|
|
Cidec
|
cell death-inducing DFFA-like effector c
|
|
chr15_-_90880241
|
11.597
|
NM_001109040
NM_001109041
NM_001109042
NM_016705
|
Kif21a
|
kinesin family member 21A
|
|
chr11_-_83880574
|
11.381
|
|
Dusp14
|
dual specificity phosphatase 14
|
|
chr15_-_75844738
|
11.339
|
NM_134087
|
Fam83h
|
family with sequence similarity 83, member H
|
|
chr12_-_105101743
|
11.302
|
|
Serpina1a
Serpina1b
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A
serine (or cysteine) preptidase inhibitor, clade A, member 1B
serine (or cysteine) peptidase inhibitor, clade A, member 1C
|
|
chr12_-_104976324
|
11.202
|
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chrX_+_160707362
|
11.104
|
NM_027153
|
Pir
|
pirin
|
|
chr14_+_31692725
|
11.086
|
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr1_+_75233043
|
10.958
|
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr12_-_105101825
|
10.953
|
NM_009243
|
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A
|
|
chr6_-_3918353
|
10.923
|
NM_009364
|
Tfpi2
|
tissue factor pathway inhibitor 2
|
|
chr15_+_54402988
|
10.812
|
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr11_-_69828831
|
10.790
|
NM_017366
|
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain
|
|
chr17_-_45729199
|
10.744
|
NM_022880
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr12_-_104976382
|
10.741
|
NM_009244
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chr9_+_78039765
|
10.672
|
NM_010357
|
Gsta4
|
glutathione S-transferase, alpha 4
|
|
chr5_-_66472208
|
10.290
|
|
Rbm47
|
RNA binding motif protein 47
|
|
chr2_+_91097485
|
10.245
|
NM_028733
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3
|
|
chr10_+_18564932
|
10.206
|
|
Perp
|
PERP, TP53 apoptosis effector
|
|
chr2_+_32483909
|
9.843
|
NM_001198792
|
Ak1
|
adenylate kinase 1
|
|
chr15_-_83002612
|
9.833
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr1_+_75233276
|
9.814
|
NM_001159885
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr1_+_75233046
|
9.806
|
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr17_+_88036047
|
9.651
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr15_+_54402914
|
9.515
|
NM_178920
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr3_-_10208575
|
9.336
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_-_148462649
|
9.063
|
NM_001114322
NM_028069
|
Cdhr5
|
cadherin-related family member 5
|
|
chr12_-_105143089
|
9.059
|
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C
|
|
chr12_-_105066203
|
8.807
|
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chr9_+_62705890
|
8.661
|
NM_001102468
NM_138304
|
Calml4
|
calmodulin-like 4
|
|
chr3_-_88307024
|
8.597
|
|
Lmna
|
lamin A
|
|
chr10_+_23614493
|
8.537
|
NM_011704
|
Vnn1
|
vanin 1
|
|
chr17_+_88035996
|
8.456
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr4_-_53156928
|
8.429
|
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr17_+_88036128
|
8.407
|
|
Epcam
|
epithelial cell adhesion molecule
|
|
chr12_-_105143108
|
8.372
|
NM_009245
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C
|
|
chr19_+_6298046
|
8.275
|
|
Ehd1
|
EH-domain containing 1
|
|
chr1_-_31006599
|
8.248
|
NM_011200
|
Ptp4a1
|
protein tyrosine phosphatase 4a1
|
|
chr2_+_151931424
|
8.200
|
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae)
|
|
chr7_-_30033483
|
8.191
|
NM_028179
|
2200002D01Rik
|
RIKEN cDNA 2200002D01 gene
|
|
chr19_-_40476799
|
8.190
|
NM_001034964
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr10_+_12810593
|
8.139
|
NM_009538
|
Plagl1
|
pleiomorphic adenoma gene-like 1
|
|
chr1_-_182123596
|
8.068
|
NM_001163290
|
Adck3
|
aarF domain containing kinase 3
|
|
chr1_-_155033576
|
8.060
|
NM_008485
|
Lamc2
|
laminin, gamma 2
|
|
chr16_-_44558929
|
8.018
|
NM_172506
|
Boc
|
biregional cell adhesion molecule-related/down-regulated by oncogenes (Cdon) binding protein
|
|
chr2_+_131735663
|
7.977
|
NM_011170
|
Prnp
|
prion protein
|
|
chr7_-_29692382
|
7.975
|
|
Actn4
|
actinin alpha 4
|
|
chr11_+_78002249
|
7.891
|
NM_033475
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr18_-_35114005
|
7.819
|
|
Hspa9
|
heat shock protein 9
|
|
chr12_-_105011795
|
7.728
|
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D
|
|
chr6_+_17231172
|
7.688
|
|
Cav2
|
caveolin 2
|
|
chr14_-_56377206
|
7.659
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr1_+_75232981
|
7.613
|
NM_001159883
NM_001159884
NM_020266
NM_178055
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr2_-_38499436
|
7.604
|
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7
|
|
chr2_+_151398367
|
7.597
|
NM_145535
|
Sdcbp2
|
syndecan binding protein (syntenin) 2
|
|
chr1_+_75233130
|
7.534
|
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chrX_+_53984926
|
7.527
|
NM_010211
|
Fhl1
|
four and a half LIM domains 1
|
|
chr14_-_56377260
|
7.486
|
NM_009894
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr9_+_108241599
|
7.471
|
NM_008160
|
Gpx1
|
glutathione peroxidase 1
|
|
chr9_+_72596925
|
7.452
|
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr19_-_4037895
|
7.428
|
NM_013541
|
Gstp1
|
glutathione S-transferase, pi 1
|
|
chr12_-_105011767
|
7.407
|
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D
|
|
chr9_+_78463352
|
7.393
|
NM_153098
|
Cd109
|
CD109 antigen
|
|
chr12_+_16901671
|
7.202
|
NM_009072
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2
|
|
chrX_+_21292025
|
7.192
|
NM_020049
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14
|
|
chr14_-_56377231
|
7.180
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr2_+_157104738
|
7.154
|
NM_019642
|
Rpn2
|
ribophorin II
|
|
chr2_-_38499375
|
7.139
|
NM_011187
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7
|
|
chr18_+_5046584
|
7.078
|
NM_153153
|
Svil
|
supervillin
|
|
chr7_-_30980042
|
6.953
|
NM_009795
|
Capns1
|
calpain, small subunit 1
|
|
chr14_-_56377278
|
6.891
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr3_+_137880777
|
6.890
|
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr11_-_116668593
|
6.719
|
|
|
|
|
chr14_-_35401845
|
6.612
|
NM_001039071
NM_001039072
NM_001039073
NM_001039075
NM_001039076
NM_011918
|
Ldb3
|
LIM domain binding 3
|
|
chrX_+_130442964
|
6.607
|
NM_001083895
NM_026838
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2
|
|
chr10_+_128370814
|
6.517
|
NM_008398
|
Itga7
|
integrin alpha 7
|
|
chr2_+_160706405
|
6.490
|
NM_022883
|
Lpin3
|
lipin 3
|
|
chr3_-_89151364
|
6.437
|
|
Adam15
|
a disintegrin and metallopeptidase domain 15 (metargidin)
|
|
chr3_+_137880725
|
6.415
|
NM_009626
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide
|
|
chr17_-_25891264
|
6.406
|
NM_018857
|
Msln
|
mesothelin
|
|
chr7_-_140179678
|
6.331
|
|
Ctbp2
|
C-terminal binding protein 2
|
|
chr18_-_35114000
|
6.274
|
|
Hspa9
|
heat shock protein 9
|
|
chr3_+_107698740
|
6.233
|
NM_010360
|
Gstm5
|
glutathione S-transferase, mu 5
|
|
chr8_-_110112368
|
6.215
|
NM_010817
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7
|
|
chr1_-_184447549
|
6.205
|
NM_009794
|
Capn2
|
calpain 2
|
|
chr4_-_58925575
|
6.156
|
NM_172381
|
AI314180
|
expressed sequence AI314180
|
|
chr12_-_105011807
|
6.114
|
NM_009246
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D
|
|
chr4_-_155148591
|
6.104
|
NM_147776
|
Vwa1
|
von Willebrand factor A domain containing 1
|
|
chr9_+_78463569
|
6.100
|
|
Cd109
|
CD109 antigen
|
|
chr5_-_114250360
|
6.035
|
NM_146162
|
Tmem119
|
transmembrane protein 119
|
|
chr1_+_75233028
|
6.034
|
|
Dnajb2
|
DnaJ (Hsp40) homolog, subfamily B, member 2
|
|
chr9_+_78039786
|
5.953
|
|
Gsta4
|
glutathione S-transferase, alpha 4
|
|
chr13_-_96388412
|
5.937
|
|
F2r
|
coagulation factor II (thrombin) receptor
|
|
chrX_+_160811458
|
5.909
|
NM_010216
|
Figf
|
c-fos induced growth factor
|
|
chr2_-_25079684
|
5.883
|
|
Tubb2c
|
tubulin, beta 2C
|
|
chr5_-_145900858
|
5.880
|
|
Pdap1
|
PDGFA associated protein 1
|
|
chr4_-_140221801
|
5.850
|
NM_001112722
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like
|
|
chr8_+_12984287
|
5.804
|
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chrX_+_12858142
|
5.770
|
NM_010028
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr13_-_19711409
|
5.714
|
NM_134065
|
Epdr1
|
ependymin related protein 1 (zebrafish)
|
|
chr2_-_110203138
|
5.701
|
NM_026271
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr2_+_151931465
|
5.690
|
NM_029688
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae)
|
|
chr11_-_50024275
|
5.669
|
NM_011018
|
Sqstm1
|
sequestosome 1
|
|
chr14_-_37781943
|
5.619
|
NM_027045
|
Gcap14
|
granule cell antiserum positive 14
|
|
chr5_+_101275148
|
5.599
|
NM_172715
|
Agpat9
|
1-acylglycerol-3-phosphate O-acyltransferase 9
|
|
chr18_+_35278553
|
5.554
|
NM_009818
|
Ctnna1
|
catenin (cadherin associated protein), alpha 1
|
|
chr1_+_58086831
|
5.535
|
|
Aox1
|
aldehyde oxidase 1
|
|
chr11_+_94189303
|
5.513
|
NM_027799
NM_146024
|
Ankrd40
|
ankyrin repeat domain 40
|
|
chr13_+_30751950
|
5.511
|
NM_134068
NM_001037955
|
Dusp22
|
dual specificity phosphatase 22
|
|
chr6_+_48854984
|
5.502
|
NM_001161621
NM_001161622
|
Abp1
|
amiloride binding protein 1 (amine oxidase, copper-containing)
|
|
chr18_-_35786880
|
5.475
|
NM_011397
|
Slc23a1
|
solute carrier family 23 (nucleobase transporters), member 1
|
|
chr3_+_79689897
|
5.417
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr4_+_128782873
|
5.343
|
|
Tmem54
|
transmembrane protein 54
|
|
chr7_-_26573483
|
5.311
|
NM_001190974
NM_001190975
NM_009465
|
Axl
|
AXL receptor tyrosine kinase
|
|
chr18_-_35113995
|
5.288
|
|
Hspa9
|
heat shock protein 9
|
|
chr17_+_35187325
|
5.259
|
|
Clic1
|
chloride intracellular channel 1
|
|
chr10_-_80363493
|
5.253
|
|
Timm13
|
translocase of inner mitochondrial membrane 13 homolog (yeast)
|
|
chr18_+_35278585
|
5.244
|
|
Ctnna1
|
catenin (cadherin associated protein), alpha 1
|
|
chrX_-_163647057
|
5.243
|
|
Tmsb4x
|
thymosin, beta 4, X chromosome
|
|
chr2_+_157105002
|
5.231
|
|
Rpn2
|
ribophorin II
|
|
chr9_-_45747000
|
5.198
|
|
Sidt2
|
SID1 transmembrane family, member 2
|
|
chr3_-_84063605
|
5.169
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr18_-_35114004
|
5.141
|
NM_010481
|
Hspa9
|
heat shock protein 9
|
|
chr4_+_128782841
|
5.085
|
|
Tmem54
|
transmembrane protein 54
|
|
chr14_-_70577478
|
5.031
|
NM_145978
|
Pdlim2
|
PDZ and LIM domain 2
|
|
chr4_+_128782859
|
4.976
|
NM_025452
|
Tmem54
|
transmembrane protein 54
|
|
chr15_-_83002561
|
4.962
|
|
Cyb5r3
|
cytochrome b5 reductase 3
|
|
chr14_-_37781912
|
4.934
|
|
Gcap14
|
granule cell antiserum positive 14
|
|
chr14_-_35401654
|
4.842
|
NM_001039074
|
Ldb3
|
LIM domain binding 3
|
|
chr8_+_96696512
|
4.826
|
NM_008630
|
Mt2
|
metallothionein 2
|
|
chr9_+_106350967
|
4.821
|
NM_029631
|
Abhd14b
|
abhydrolase domain containing 14b
|
|
chr8_+_121961044
|
4.815
|
NM_027950
|
Osgin1
|
oxidative stress induced growth inhibitor 1
|
|
chr3_-_84024766
|
4.808
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr6_+_42299820
|
4.755
|
NM_011777
|
Zyx
|
zyxin
|
|
chr10_-_60609031
|
4.752
|
|
Sgpl1
|
sphingosine phosphate lyase 1
|
|
chr16_+_20651724
|
4.722
|
NM_134101
|
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2
|
|
chr14_-_37781899
|
4.704
|
|
Gcap14
|
granule cell antiserum positive 14
|
|
chr6_+_17231334
|
4.644
|
NM_016900
|
Cav2
|
caveolin 2
|
|
chr11_+_94189524
|
4.629
|
|
Ankrd40
|
ankyrin repeat domain 40
|
|
chr4_-_117963950
|
4.609
|
NM_011213
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F
|