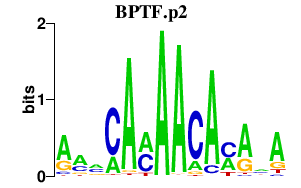
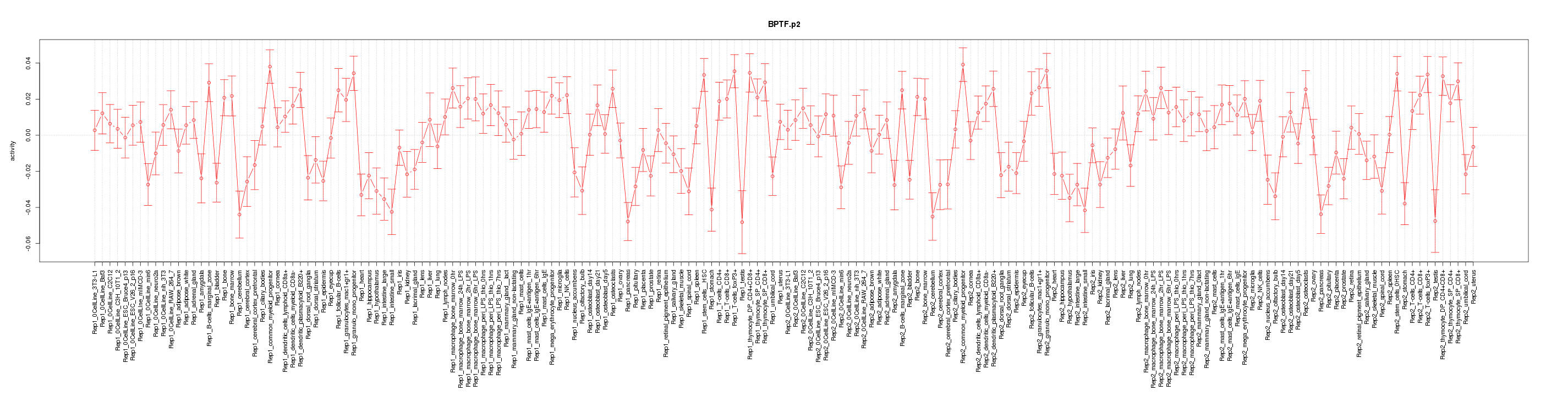
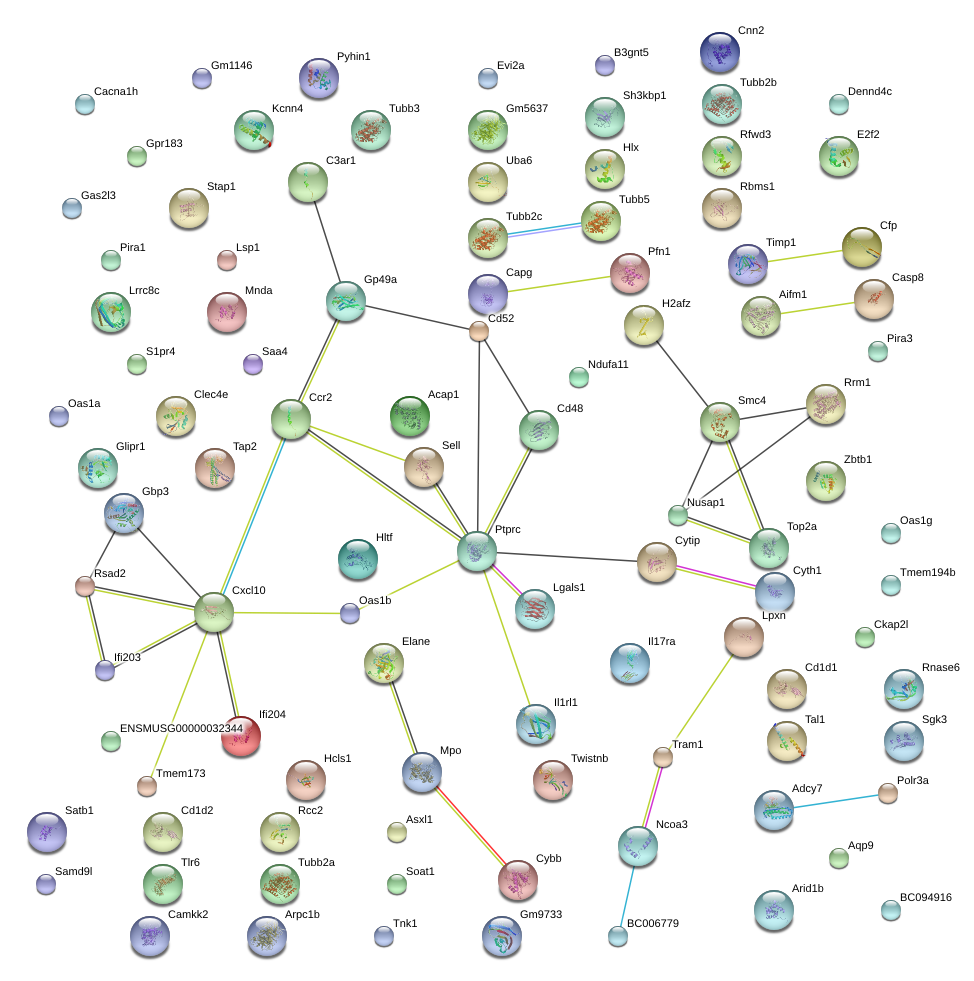
|
chr19_+_12873115
|
40.643
|
|
Lpxn
|
leupaxin
|
|
chr5_+_145888588
|
39.489
|
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B
|
|
chr11_+_83037063
|
38.808
|
|
AI662270
|
expressed sequence AI662270
|
|
chr8_+_90812964
|
34.604
|
NM_001037723
|
Adcy7
|
adenylate cyclase 7
|
|
chr2_-_60589059
|
31.980
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr19_+_12873063
|
31.863
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr5_-_86539474
|
30.446
|
|
Uba6
|
ubiquitin-like modifier activating enzyme 6
|
|
chr6_-_122806122
|
30.343
|
NM_009779
|
C3ar1
|
complement component 3a receptor 1
|
|
chr1_+_175560938
|
29.700
|
NM_175026
|
Pyhin1
|
pyrin and HIN domain family, member 1
|
|
chr4_-_133650937
|
29.655
|
NM_013706
|
Cd52
|
CD52 antigen
|
|
chr14_+_51748742
|
28.344
|
NM_030098
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr11_+_87607531
|
28.064
|
|
Mpo
|
myeloperoxidase
|
|
chr12_-_27129497
|
26.710
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr6_-_122806020
|
26.679
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr6_-_122806042
|
26.354
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr5_-_121357467
|
26.312
|
NM_145211
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A
|
|
chr1_+_165992203
|
26.145
|
NM_001164059
NM_011346
|
Sell
|
selectin, lymphocyte
|
|
chr10_+_79457716
|
25.975
|
|
Cnn2
|
calponin 2
|
|
chr1_-_175805812
|
24.695
|
|
Mndal
|
myeloid nuclear differentiation antigen like
|
|
chr1_-_140071818
|
23.754
|
NM_001111316
NM_011210
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chr1_+_40497472
|
23.728
|
NM_001025602
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr7_+_149646740
|
22.149
|
NM_001136071
|
Lsp1
|
lymphocyte specific 1
|
|
chr7_+_149646716
|
21.058
|
|
Lsp1
|
lymphocyte specific 1
|
|
chr6_+_72494474
|
20.614
|
|
Capg
|
capping protein (actin filament), gelsolin-like
|
|
chr11_+_87569075
|
20.354
|
|
A430104N18Rik
|
RIKEN cDNA A430104N18 gene
|
|
chrX_-_9014351
|
20.227
|
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr2_-_180976728
|
19.450
|
NM_183162
|
BC006779
|
cDNA sequence BC006779
|
|
chr6_+_120413200
|
18.121
|
NM_008359
|
Il17ra
|
interleukin 17 receptor A
|
|
chr2_-_60590923
|
17.619
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr18_-_35900005
|
17.467
|
NM_028261
|
Tmem173
|
transmembrane protein 173
|
|
chr1_-_158404299
|
17.069
|
NM_009230
|
Soat1
|
sterol O-acyltransferase 1
|
|
chr16_+_36935029
|
16.709
|
NM_008225
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1
|
|
chrX_-_20508584
|
16.279
|
NM_008823
|
Cfp
|
complement factor properdin
|
|
chr5_-_123183537
|
16.133
|
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr17_+_34341467
|
15.573
|
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr9_+_124016886
|
15.304
|
NM_009915
|
Ccr2
|
chemokine (C-C motif) receptor 2
|
|
chr3_-_86803173
|
15.223
|
NM_007639
|
Cd1d1
|
CD1d1 antigen
|
|
chr7_+_16633755
|
15.156
|
|
2010004M13Rik
|
RIKEN cDNA 2010004M13 gene
|
|
chr14_-_122364309
|
15.001
|
NM_183031
|
Gpr183
|
G protein-coupled receptor 183
|
|
chr6_+_72494378
|
14.348
|
NM_007599
|
Capg
|
capping protein (actin filament), gelsolin-like
|
|
chr15_+_78757171
|
14.270
|
|
Lgals1
|
lectin, galactose binding, soluble 1
|
|
chr17_+_34341417
|
14.187
|
NM_011530
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr12_+_34114475
|
13.625
|
NM_172253
|
Twistnb
|
TWIST neighbor
|
|
chr6_-_123239765
|
13.375
|
|
Clec4e
|
C-type lectin domain family 4, member e
|
|
chr7_+_111392968
|
13.291
|
NM_030684
|
Trim34a
|
tripartite motif-containing 34A
|
|
chr11_-_69709003
|
13.221
|
NM_153788
|
Acap1
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1
|
|
chr5_-_92776160
|
13.106
|
|
Cxcl10
|
chemokine (C-X-C motif) ligand 10
|
|
chr14_-_25306261
|
13.015
|
NM_001081247
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A
|
|
chr10_-_88875526
|
12.906
|
|
Gas2l3
|
growth arrest-specific 2 like 3
|
|
chr3_+_89904315
|
12.681
|
|
|
|
|
chr12_-_66273564
|
12.623
|
NM_172578
|
Mis18bp1
|
MIS18 binding protein 1
|
|
chr5_+_86500770
|
12.585
|
NM_019992
|
Stap1
|
signal transducing adaptor family member 1
|
|
chr9_-_78290953
|
12.552
|
NM_173386
|
E330016A19Rik
|
RIKEN cDNA E330016A19 gene
|
|
chr12_+_77471173
|
12.351
|
NM_178744
|
Zbtb1
|
zinc finger and BTB domain containing 1
|
|
chr1_+_58859345
|
12.226
|
NM_001080126
|
Casp8
|
caspase 8
|
|
chr6_+_116123984
|
12.196
|
|
Gm8203
|
predicted pseudogene 8203
|
|
chr2_-_58012447
|
12.038
|
NM_139200
|
Cytip
|
cytohesin 1 interacting protein
|
|
chr1_-_140071630
|
11.900
|
|
Ptprc
|
protein tyrosine phosphatase, receptor type, C
|
|
chr3_+_142223490
|
11.886
|
|
Gbp3
|
guanylate binding protein 3
|
|
chr10_-_111434231
|
11.712
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr10_+_51200463
|
11.671
|
NM_008147
|
Gp49a
|
glycoprotein 49 A
|
|
chr17_-_35971652
|
11.622
|
|
Tubb5
|
tubulin, beta 5
|
|
chr17_+_5343877
|
11.566
|
|
Arid1b
|
AT rich interactive domain 1B (SWI-like)
|
|
chr1_+_52687548
|
11.409
|
NM_001142647
|
Tmem194b
|
transmembrane protein 194B
|
|
chr2_-_129094163
|
11.355
|
|
Ckap2l
|
cytoskeleton associated protein 2-like
|
|
chr5_+_106039499
|
11.312
|
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C
|
|
chr10_+_126504125
|
11.263
|
|
|
|
|
chr19_+_11566527
|
11.242
|
NM_001164202
|
Gm8369
|
predicted gene 8369
|
|
chr11_-_79343891
|
11.236
|
|
Evi2a
|
ecotropic viral integration site 2a
|
|
chr1_+_173612173
|
10.991
|
NM_007649
|
Cd48
|
CD48 antigen
|
|
chr9_-_71011053
|
10.881
|
NM_022026
|
Aqp9
|
aquaporin 9
|
|
chr11_-_98859189
|
10.684
|
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr6_-_3349415
|
10.594
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr8_-_113824073
|
10.579
|
NM_146218
|
Rfwd3
|
ring finger and WD repeat domain 3
|
|
chrX_+_20451640
|
10.484
|
|
Timp1
|
tissue inhibitor of metalloproteinase 1
|
|
chr11_-_70465629
|
10.400
|
|
Pfn1
|
profilin 1
|
|
chr2_+_153171730
|
10.274
|
NM_001039939
|
Asxl1
|
additional sex combs like 1 (Drosophila)
|
|
chr6_-_98877291
|
10.250
|
|
|
|
|
chr7_+_25155279
|
10.249
|
NM_001163510
NM_008433
|
Kcnn4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4
|
|
chr4_+_128688217
|
10.239
|
|
|
|
|
chr10_-_80962881
|
10.236
|
NM_010102
|
S1pr4
|
sphingosine-1-phosphate receptor 4
|
|
chr1_+_9788206
|
10.222
|
NM_133220
NM_177547
|
Sgk3
|
serum/glucocorticoid regulated kinase 3
|
|
chr4_+_114732112
|
10.164
|
NM_011527
|
Tal1
|
T-cell acute lymphocytic leukemia 1
|
|
chr4_+_86394436
|
10.038
|
NM_001081014
NM_184088
|
Dennd4c
|
DENN/MADD domain containing 4C
|
|
chrX_+_155970637
|
10.034
|
|
A830080D01Rik
|
RIKEN cDNA A830080D01 gene
|
|
chr7_+_4225744
|
10.033
|
NM_011093
NM_008848
|
Pira6
|
paired-Ig-like receptor A6
|
|
chr4_+_140258090
|
9.975
|
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr5_-_65351226
|
9.960
|
NM_011604
|
Tlr6
|
toll-like receptor 6
|
|
chr7_+_109590208
|
9.876
|
NM_009103
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr17_+_34052312
|
9.870
|
|
|
|
|
chr12_-_27141303
|
9.844
|
NM_021384
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr2_+_80193710
|
9.834
|
|
|
|
|
chr7_-_25873560
|
9.834
|
|
|
|
|
chr4_+_135750500
|
9.731
|
|
E2f2
|
E2F transcription factor 2
|
|
chr1_-_186556046
|
9.673
|
|
Hlx
|
H2.0-like homeobox
|
|
chrX_-_45866446
|
9.657
|
NM_012019
|
Aifm1
|
apoptosis-inducing factor, mitochondrion-associated 1
|
|
chr10_+_79348991
|
9.648
|
NM_015779
|
Elane
|
elastase, neutrophil expressed
|
|
chrX_+_155970593
|
9.582
|
NM_001033472
|
A830080D01Rik
|
RIKEN cDNA A830080D01 gene
|
|
chr1_-_13579802
|
9.486
|
|
Tram1
|
translocating chain-associating membrane protein 1
|
|
chr16_+_19760300
|
9.481
|
NM_001159407
NM_001159408
NM_054052
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr3_+_68808879
|
9.451
|
NM_133786
|
Smc4
|
structural maintenance of chromosomes 4
|
|
chr6_-_123239886
|
9.348
|
NM_019948
|
Clec4e
|
C-type lectin domain family 4, member e
|
|
chr15_-_72926936
|
9.333
|
|
1110029L17Rik
|
RIKEN cDNA 1110029L17 gene
|
|
chr14_-_25306175
|
9.265
|
|
Polr3a
|
polymerase (RNA) III (DNA directed) polypeptide A
|
|
chrX_+_156065141
|
9.240
|
NM_001135727
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr17_-_51951378
|
9.226
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr3_+_20008792
|
9.219
|
|
Hltf
|
helicase-like transcription factor
|
|
chr2_+_119444033
|
9.175
|
NM_001042652
NM_133851
|
Nusap1
|
nucleolar and spindle associated protein 1
|
|
chr8_+_47702617
|
8.910
|
|
Casp3
|
caspase 3
|
|
chr12_-_32893074
|
8.858
|
NM_001146201
NM_020272
NM_001146200
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr11_-_102180258
|
8.855
|
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr12_+_119082333
|
8.812
|
NM_146040
|
Cdca7l
|
cell division cycle associated 7 like
|
|
chr6_+_136467315
|
8.778
|
NM_019426
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr1_-_175465858
|
8.760
|
|
BC094916
|
cDNA sequence BC094916
|
|
chr11_-_54601093
|
8.702
|
|
Cdc42se2
|
CDC42 small effector 2
|
|
chr17_+_34375847
|
8.615
|
NM_010389
|
H2-Ob
|
histocompatibility 2, O region beta locus
|
|
chr6_+_38542722
|
8.615
|
|
Luc7l2
|
LUC7-like 2 (S. cerevisiae)
|
|
chr13_+_55828701
|
8.558
|
NM_028281
|
Pcbd2
|
pterin 4 alpha carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2
|
|
chr9_+_123429291
|
8.510
|
|
Limd1
|
LIM domains containing 1
|
|
chr17_-_46290499
|
8.478
|
NM_025649
|
Mad2l1bp
|
MAD2L1 binding protein
|
|
chr10_-_26093761
|
8.452
|
|
L3mbtl3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr10_-_90633199
|
8.447
|
|
Tmpo
|
thymopoietin
|
|
chr16_+_49913305
|
8.347
|
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer)
|
|
chr2_+_150735306
|
8.339
|
NM_001163476
NM_027014
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr13_-_56280161
|
8.315
|
NM_001168615
NM_145976
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B
|
|
chr15_+_83393975
|
8.232
|
NM_009775
|
Tspo
|
translocator protein
|
|
chr7_+_65913571
|
8.211
|
NM_009728
|
Atp10a
|
ATPase, class V, type 10A
|
|
chr2_+_103809709
|
8.202
|
NM_001142336
|
Lmo2
|
LIM domain only 2
|
|
chr7_-_56136965
|
8.174
|
|
E2f8
|
E2F transcription factor 8
|
|
chr11_-_106640755
|
8.150
|
NM_015810
|
Polg2
|
polymerase (DNA directed), gamma 2, accessory subunit
|
|
chr8_+_90806890
|
8.071
|
NM_001109756
|
Adcy7
|
adenylate cyclase 7
|
|
chr13_-_67710581
|
8.043
|
|
AA987161
|
expressed sequence AA987161
|
|
chr15_+_78427476
|
8.018
|
NM_028195
|
Cyth4
|
cytohesin 4
|
|
chr16_-_75909440
|
7.959
|
NM_023380
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1
|
|
chr1_-_173582969
|
7.939
|
NM_144539
|
Slamf7
|
SLAM family member 7
|
|
chr1_+_173612132
|
7.934
|
|
Cd48
|
CD48 antigen
|
|
chr1_+_132762248
|
7.860
|
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr2_+_129442079
|
7.850
|
|
Sirpa
|
signal-regulatory protein alpha
|
|
chr18_-_14131046
|
7.783
|
NM_145492
|
Zfp521
|
zinc finger protein 521
|
|
chr10_-_58904445
|
7.781
|
NM_010959
|
Oit3
|
oncoprotein induced transcript 3
|
|
chr6_-_38238095
|
7.717
|
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like
|
|
chr11_+_81859212
|
7.712
|
NM_013654
|
Ccl7
|
chemokine (C-C motif) ligand 7
|
|
chr7_-_125629996
|
7.710
|
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chrX_-_103163796
|
7.697
|
|
Magt1
|
magnesium transporter 1
|
|
chr12_-_119474010
|
7.611
|
|
Sp4
|
trans-acting transcription factor 4
|
|
chr12_-_27141277
|
7.609
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chrX_+_50265390
|
7.603
|
NM_027642
|
Phf6
|
PHD finger protein 6
|
|
chr11_-_102180282
|
7.567
|
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr2_-_126719808
|
7.530
|
|
2010106G01Rik
|
RIKEN cDNA 2010106G01 gene
|
|
chr2_-_51828440
|
7.512
|
|
Nmi
|
N-myc (and STAT) interactor
|
|
chr6_-_30340952
|
7.498
|
NM_172735
|
Zc3hc1
|
zinc finger, C3HC type 1
|
|
chr10_-_12684064
|
7.496
|
NM_001163590
NM_001163591
NM_029075
|
Stx11
|
syntaxin 11
|
|
chrX_+_83115611
|
7.458
|
NM_001163539
|
5430427O19Rik
|
RIKEN cDNA 5430427O19 gene
|
|
chr5_+_93602126
|
7.423
|
|
Sept11
|
septin 11
|
|
chr4_+_140703847
|
7.414
|
|
Fbxo42
|
F-box protein 42
|
|
chr13_-_74832039
|
7.399
|
|
Cast
|
calpastatin
|
|
chr17_-_41071432
|
7.392
|
NM_031863
|
Cenpq
|
centromere protein Q
|
|
chr5_+_138628733
|
7.386
|
NM_178612
|
Cnpy4
|
canopy 4 homolog (zebrafish)
|
|
chrX_-_154036557
|
7.368
|
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr10_-_90633803
|
7.342
|
|
Tmpo
|
thymopoietin
|
|
chr2_-_113688771
|
7.338
|
NM_181416
|
Arhgap11a
|
Rho GTPase activating protein 11A
|
|
chr17_-_46290473
|
7.328
|
|
Mad2l1bp
|
MAD2L1 binding protein
|
|
chr1_-_180267913
|
7.283
|
NM_016805
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr7_-_3796619
|
7.273
|
|
Pira1
Pira2
Lilrb3
|
paired-Ig-like receptor A1
paired-Ig-like receptor A2
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
|
|
chr5_+_124199713
|
7.232
|
NM_001042421
|
Kntc1
|
kinetochore associated 1
|
|
chr6_+_137703029
|
7.228
|
NM_172733
|
Dera
|
2-deoxyribose-5-phosphate aldolase homolog (C. elegans)
|
|
chr7_-_3849680
|
7.171
|
NM_001166672
NM_011088
NM_011091
NM_011094
|
Gm14548
Pira11
Pira4
Pira5
Pira7
Lilrb3
|
predicted gene 14548
paired-Ig-like receptor A11
paired-Ig-like receptor A4
paired-Ig-like receptor A5
paired-Ig-like receptor A7
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
|
|
chr7_-_111501738
|
7.153
|
NM_175677
NM_001146007
|
9230105E10Rik
|
RIKEN cDNA 9230105E10 gene
|
|
chr2_-_6466116
|
7.113
|
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr5_+_115346942
|
7.113
|
NM_011854
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr4_-_6202747
|
7.055
|
NM_007824
|
Cyp7a1
|
cytochrome P450, family 7, subfamily a, polypeptide 1
|
|
chr6_+_122576437
|
7.042
|
NM_139218
|
Dppa3
|
developmental pluripotency-associated 3
|
|
chr8_-_40727989
|
7.025
|
NM_001113326
NM_031195
|
Msr1
|
macrophage scavenger receptor 1
|
|
chr9_-_44334460
|
7.017
|
NM_007551
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5
|
|
chr17_-_35971913
|
7.004
|
|
Tubb5
|
tubulin, beta 5
|
|
chr11_-_53520771
|
6.923
|
|
Rad50
|
RAD50 homolog (S. cerevisiae)
|
|
chr6_-_30341017
|
6.918
|
|
Zc3hc1
|
zinc finger, C3HC type 1
|
|
chr6_-_129187965
|
6.893
|
|
2310001H17Rik
|
RIKEN cDNA 2310001H17 gene
|
|
chr6_+_48689045
|
6.874
|
NM_008376
NM_175860
|
Gimap1
|
GTPase, IMAP family member 1
|
|
chr17_-_71199129
|
6.859
|
NM_001164076
|
Tgif1
|
TGFB-induced factor homeobox 1
|
|
chr11_-_102180374
|
6.827
|
NM_011551
|
Ubtf
|
upstream binding transcription factor, RNA polymerase I
|
|
chr8_+_107694536
|
6.802
|
NM_001161456
NM_001161457
NM_001161458
NM_022309
|
Cbfb
|
core binding factor beta
|
|
chr13_+_33057312
|
6.782
|
NM_011454
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b
|
|
chr7_-_87857062
|
6.781
|
|
|
|
|
chr15_-_80390899
|
6.768
|
NM_001163189
|
Enthd1
|
ENTH domain containing 1
|
|
chr5_-_66000506
|
6.726
|
|
B230308N11Rik
|
RIKEN cDNA B230308N11 gene
|
|
chr12_-_80108263
|
6.723
|
NM_178745
|
Tmem229b
|
transmembrane protein 229B
|
|
chr10_+_40290141
|
6.714
|
NM_027572
|
Slc22a16
|
solute carrier family 22 (organic cation transporter), member 16
|
|
chr16_+_23224955
|
6.666
|
|
St6gal1
|
beta galactoside alpha 2,6 sialyltransferase 1
|
|
chr7_+_109614267
|
6.659
|
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr11_+_54336372
|
6.652
|
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6
|
|
chr9_+_106109125
|
6.650
|
|
Twf2
|
twinfilin, actin-binding protein, homolog 2 (Drosophila)
|
|
chr12_+_82049290
|
6.641
|
|
Srsf5
|
serine/arginine-rich splicing factor 5
|
|
chrX_-_9046362
|
6.628
|
NM_007807
|
Cybb
|
cytochrome b-245, beta polypeptide
|
|
chr2_+_112124767
|
6.571
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr1_+_87546558
|
6.540
|
NM_013673
|
Sp100
|
nuclear antigen Sp100
|
|
chr14_+_51563976
|
6.497
|
NM_013632
|
Pnp
|
purine-nucleoside phosphorylase
|
|
chr11_+_83251195
|
6.471
|
NM_025492
|
1700020L24Rik
|
RIKEN cDNA 1700020L24 gene
|
|
chr7_+_77509969
|
6.464
|
|
B130024G19Rik
|
RIKEN cDNA B130024G19 gene
|
|
chr9_-_114690887
|
6.456
|
NM_133978
|
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7
|