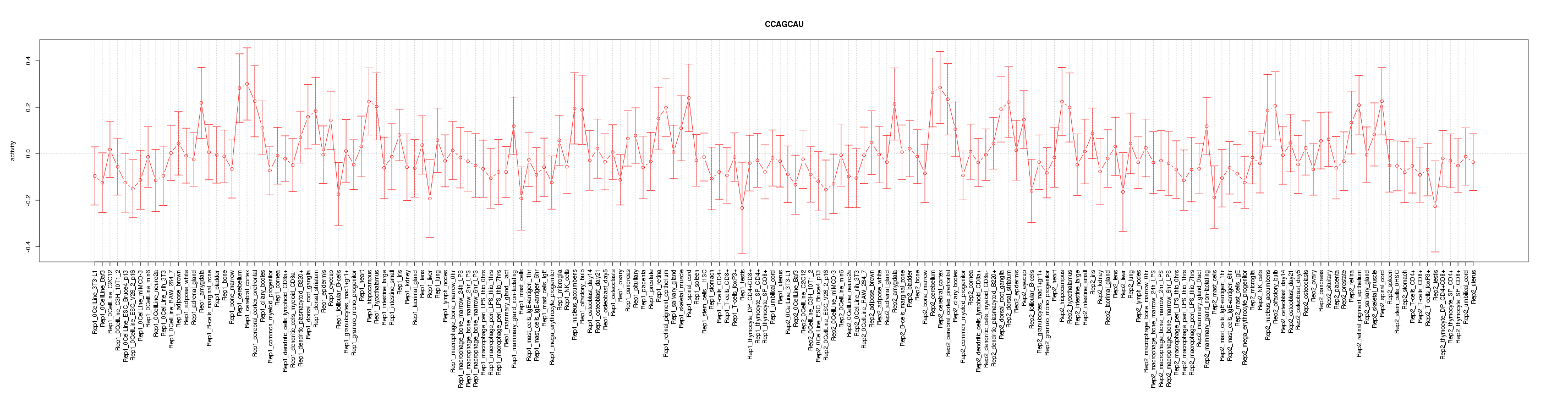
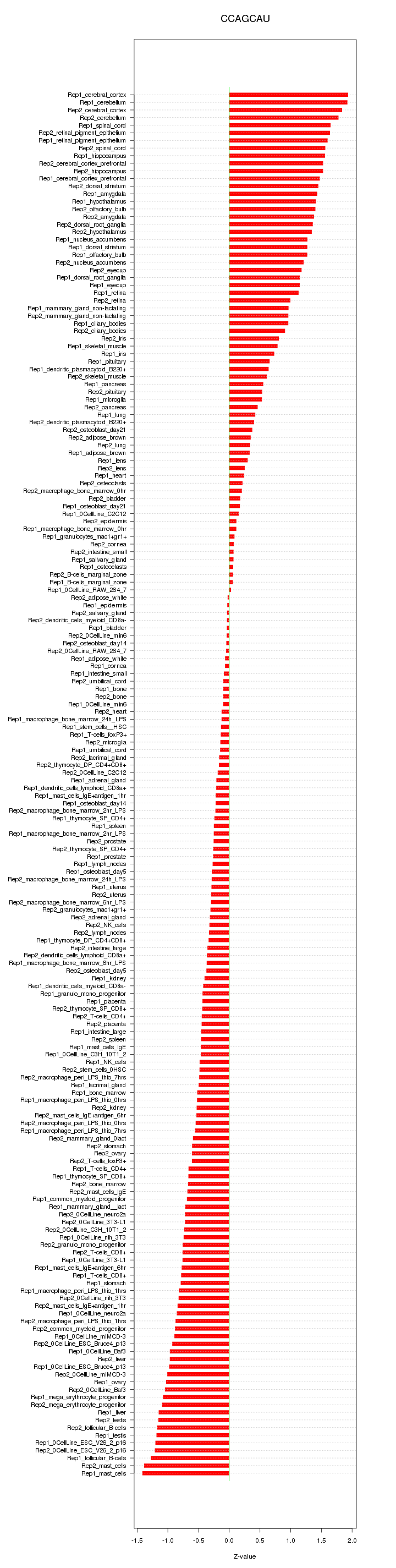
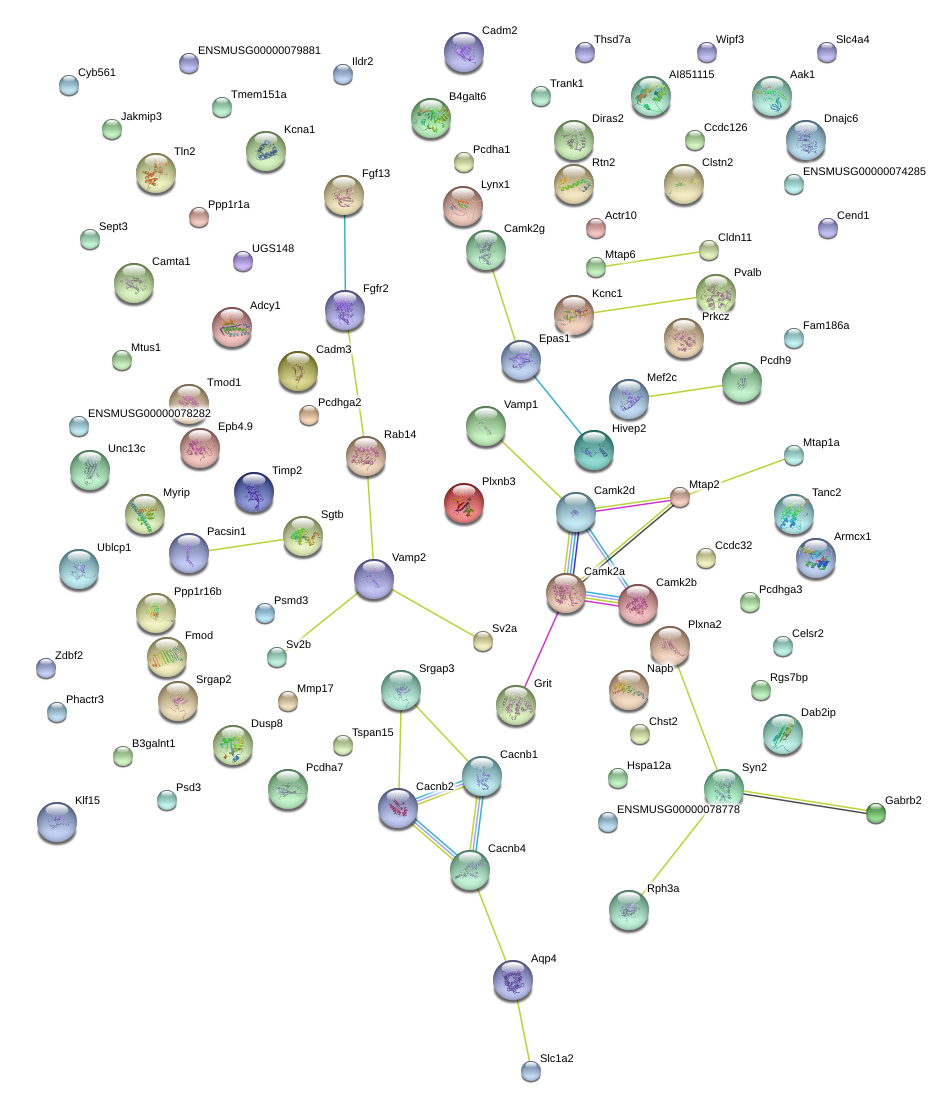
|
chr15_-_78034565
|
10.708
|
NM_013645
|
Pvalb
|
parvalbumin
|
|
chr19_-_58935470
|
7.825
|
NM_175199
|
Hspa12a
|
heat shock protein 12A
|
|
chr4_-_151235861
|
7.650
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr16_-_67621054
|
7.456
|
NM_001145977
NM_178721
|
Cadm2
|
cell adhesion molecule 2
|
|
chr9_+_32031833
|
7.398
|
NM_177379
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr13_-_105844882
|
6.866
|
NM_029879
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr8_-_70342105
|
6.571
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr6_+_86799460
|
6.457
|
NM_001040106
NM_177762
|
Aak1
|
AP2 associated kinase 1
|
|
chr11_-_70847078
|
6.187
|
NM_134022
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene
|
|
chr13_+_83643032
|
5.953
|
NM_001170537
NM_025282
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr18_+_37906739
|
5.933
|
NM_033593
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr18_+_37849488
|
5.926
|
NM_033575
|
Pcdhgb2
|
protocadherin gamma subfamily B, 2
|
|
chr18_+_37915375
|
5.809
|
NM_033594
|
Pcdhga11
|
protocadherin gamma subfamily A, 11
|
|
chr7_-_137410321
|
5.797
|
NM_010207
NM_201601
|
Fgfr2
|
fibroblast growth factor receptor 2
|
|
chr18_+_37844927
|
5.769
|
NM_033587
|
Pcdhga4
|
protocadherin gamma subfamily A, 4
|
|
chr18_+_61123204
|
5.695
|
NM_009792
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr18_+_37833939
|
5.664
|
NM_033586
|
Pcdhga3
|
protocadherin gamma subfamily A, 3
|
|
chr18_+_37828606
|
5.621
|
NM_033585
|
Pcdhga2
|
protocadherin gamma subfamily A, 2
|
|
chr18_+_37966017
|
5.615
|
NM_033581
|
Pcdhgc3
|
protocadherin gamma subfamily C, 3
|
|
chr18_+_37890662
|
5.608
|
NM_033577
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5
|
|
chr2_-_52532100
|
5.600
|
NM_001037099
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr2_-_52414072
|
5.530
|
NM_146123
|
Cacnb4
|
calcium channel, voltage-dependent, beta 4 subunit
|
|
chr18_+_37901716
|
5.455
|
NM_033578
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6
|
|
chr18_+_37885359
|
5.324
|
NM_033591
|
Pcdhga8
|
protocadherin gamma subfamily A, 8
|
|
chr15_+_82105322
|
5.219
|
NM_011889
|
Sept3
|
septin 3
|
|
chr11_+_68902025
|
5.153
|
NM_009497
|
Vamp2
|
vesicle-associated membrane protein 2
|
|
chr18_+_37839910
|
5.122
|
NM_033574
|
Pcdhgb1
|
protocadherin gamma subfamily B, 1
|
|
chr18_+_37911236
|
5.090
|
NM_033579
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7
|
|
chr18_+_37896589
|
5.087
|
NM_033592
|
Pcdhga9
|
protocadherin gamma subfamily A, 9
|
|
chr15_+_87455659
|
5.050
|
NM_134096
|
Fam19a5
|
family with sequence similarity 19, member A5
|
|
chr18_+_37821598
|
4.837
|
NM_033584
|
Pcdhga1
|
protocadherin gamma subfamily A, 1
|
|
chr18_+_37925233
|
4.803
|
NM_033595
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr18_+_37874471
|
4.768
|
NM_033590
|
Pcdhga7
|
protocadherin gamma subfamily A, 7
|
|
chr18_+_37921454
|
4.724
|
NM_033580
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8
|
|
chr18_+_37979199
|
4.720
|
NM_033583
|
Pcdhgb4
Pcdhgc5
|
protocadherin gamma subfamily B, 4
protocadherin gamma subfamily C, 5
|
|
chr9_+_31923720
|
4.713
|
NM_001195632
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr2_-_144353133
|
4.685
|
NM_172859
|
6330439K17Rik
|
RIKEN cDNA 6330439K17 gene
|
|
chr18_+_37866747
|
4.615
|
NM_033589
|
Pcdhga6
|
protocadherin gamma subfamily A, 6
|
|
chr2_+_35477498
|
4.614
|
NM_001114124
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr2_-_148558110
|
4.409
|
NM_019632
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta
|
|
chr7_-_82454147
|
4.399
|
NM_153579
|
Sv2b
|
synaptic vesicle glycoprotein 2 b
|
|
chr18_+_37880036
|
4.357
|
NM_033576
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4
|
|
chr14_-_21613162
|
4.351
|
NM_001039138
NM_001039139
NM_178597
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma
|
|
chr5_+_130090042
|
4.337
|
NM_011846
|
Mmp17
|
matrix metallopeptidase 17
|
|
chr15_-_74583382
|
4.327
|
NM_011838
|
Lynx1
|
Ly6/neurotoxin 1
|
|
chr2_+_102498839
|
4.266
|
NM_001077514
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr4_-_154735351
|
4.243
|
NM_008860
|
Prkcz
|
protein kinase C, zeta
|
|
chr18_+_37854011
|
4.208
|
NM_033588
|
Pcdhga5
|
protocadherin gamma subfamily A, 5
|
|
chr14_-_71029960
|
4.140
|
NM_013514
|
Epb4.9
|
erythrocyte protein band 4.9
|
|
chr7_-_82453216
|
4.129
|
NM_001109753
|
Sv2b
|
synaptic vesicle glycoprotein 2 b
|
|
chr5_+_89316245
|
4.127
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr18_+_37974732
|
4.018
|
NM_033582
|
Pcdhgc4
|
protocadherin gamma subfamily C, 4
|
|
chr18_-_20904585
|
4.015
|
NM_019737
|
B4galt6
|
UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 6
|
|
chr2_+_35547418
|
3.972
|
NM_001001602
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr9_-_67407509
|
3.925
|
NM_001081242
|
Tln2
|
talin 2
|
|
chr8_-_70434808
|
3.900
|
NM_030263
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr4_-_154719834
|
3.890
|
NM_001039079
|
Prkcz
|
protein kinase C, zeta
|
|
chr2_+_177910053
|
3.755
|
NM_001177791
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr17_+_27792587
|
3.708
|
NM_011861
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr2_+_177853679
|
3.670
|
NM_001177789
NM_001177790
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr2_+_177927788
|
3.660
|
NM_001007154
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr11_-_44283916
|
3.630
|
NM_024475
|
Ublcp1
|
ubiquitin-like domain containing CTD phosphatase 1
|
|
chr10_+_13686174
|
3.506
|
NM_010437
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2
|
|
chr1_+_66221851
|
3.469
|
NM_001039934
NM_008632
|
Mtap2
|
microtubule-associated protein 2
|
|
chr6_-_112897259
|
3.439
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr1_+_135934035
|
3.437
|
NM_021355
|
Fmod
|
fibromodulin
|
|
chrX_-_56387565
|
3.415
|
NM_010200
|
Fgf13
|
fibroblast growth factor 13
|
|
chr4_+_101169252
|
3.302
|
NM_198412
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr7_+_19876447
|
3.288
|
NM_001025364
|
Rtn2
|
reticulon 2 (Z-band associated protein)
|
|
chr6_+_54402876
|
3.270
|
NM_001167860
NM_001167861
|
Wipf3
|
WAS/WASL interacting protein family, member 3
|
|
chr3_-_108218411
|
3.243
|
NM_001004177
NM_017392
|
Celsr2
|
cadherin, EGF LAG seven-pass G-type receptor 2 (flamingo homolog, Drosophila)
|
|
chr9_+_120213190
|
3.162
|
NM_144557
|
Myrip
|
myosin VIIA and Rab interacting protein
|
|
chr18_-_15562126
|
3.131
|
NM_009700
|
Aqp4
|
aquaporin 4
|
|
chr14_-_94287950
|
3.097
|
NM_001081377
|
Pcdh9
|
protocadherin 9
|
|
chr19_-_5085471
|
3.093
|
NM_001001885
|
Tmem151a
|
transmembrane protein 151A
|
|
chr13_+_104899869
|
3.077
|
NM_144838
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr1_+_196446006
|
3.076
|
NM_008882
|
Plxna2
|
plexin A2
|
|
chr2_+_177876635
|
3.034
|
NM_028806
|
Phactr3
|
phosphatase and actin regulator 3
|
|
chr7_+_19868009
|
3.018
|
NM_013648
|
Rtn2
|
reticulon 2 (Z-band associated protein)
|
|
chr2_-_35056582
|
3.012
|
NM_026697
|
Rab14
|
RAB14, member RAS oncogene family
|
|
chr2_+_102546576
|
2.958
|
NM_001077515
NM_011393
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr6_+_49269351
|
2.900
|
NM_175098
|
Ccdc126
|
coiled-coil domain containing 126
|
|
chr3_-_69402739
|
2.889
|
NM_020026
|
B3galnt1
|
UDP-GalNAc:betaGlcNAc beta 1,3-galactosaminyltransferase, polypeptide 1
|
|
chr1_+_63319842
|
2.870
|
NM_028673
|
Zdbf2
|
zinc finger, DBF-type containing 2
|
|
chr8_-_42140113
|
2.847
|
NM_001005864
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr9_-_73781373
|
2.839
|
NM_001081153
|
Unc13c
|
unc-13 homolog C (C. elegans)
|
|
chr1_+_168184269
|
2.832
|
NM_001164528
|
Ildr2
|
immunoglobulin-like domain containing receptor 2
|
|
chr6_+_115084919
|
2.823
|
NM_001111015
NM_013681
|
Syn2
|
synapsin II
|
|
chr10_-_61693963
|
2.823
|
NM_197996
|
Tspan15
|
tetraspanin 15
|
|
chr11_-_118216689
|
2.755
|
NM_011594
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr18_+_37089837
|
2.753
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr18_+_37120093
|
2.743
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr18_+_37098858
|
2.718
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr18_+_37179802
|
2.717
|
NM_138663
|
Pcdha12
|
protocadherin alpha 12
|
|
chrX_+_131252456
|
2.715
|
NM_001166377
NM_001166378
NM_001166379
NM_001166380
NM_030066
|
Armcx1
|
armadillo repeat containing, X-linked 1
|
|
chr2_+_121115337
|
2.711
|
NM_032393
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr7_-_148615247
|
2.694
|
NM_021316
|
Cend1
|
cell cycle exit and neuronal differentiation 1
|
|
chr18_+_37127284
|
2.679
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr7_+_146132386
|
2.664
|
NM_028708
|
Jakmip3
|
janus kinase and microtubule interacting protein 3
|
|
chr17_+_87153083
|
2.637
|
NM_010137
|
Epas1
|
endothelial PAS domain protein 1
|
|
chr5_-_121459496
|
2.611
|
NM_011286
|
Rph3a
|
rabphilin 3A
|
|
chr6_-_126595818
|
2.609
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr3_+_95985065
|
2.569
|
NM_022030
|
Sv2a
|
synaptic vesicle glycoprotein 2 a
|
|
chr6_-_12699189
|
2.543
|
NM_001164805
|
Thsd7a
|
thrombospondin, type I, domain containing 7A
|
|
chr9_-_97933533
|
2.534
|
NM_022319
|
Clstn2
|
calsyntenin 2
|
|
chr18_+_37157533
|
2.518
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chr7_-_149276174
|
2.502
|
NM_008748
|
Dusp8
|
dual specificity phosphatase 8
|
|
chr11_+_6963421
|
2.470
|
NM_009622
|
Adcy1
|
adenylate cyclase 1
|
|
chr18_+_37105758
|
2.429
|
NM_138662
|
Pcdha3
|
protocadherin alpha 3
|
|
chr15_-_103368312
|
2.419
|
NM_021391
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A
|
|
chr12_+_72038843
|
2.404
|
NM_019785
|
Actr10
|
ARP10 actin-related protein 10 homolog (S. cerevisiae)
|
|
chr2_+_121121189
|
2.363
|
NM_001173506
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr1_-_175297778
|
2.353
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chr18_+_37152024
|
2.327
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr7_+_106416854
|
2.325
|
NM_001043355
|
Mtap6
|
microtubule-associated protein 6
|
|
chr6_+_90412566
|
2.323
|
NM_023184
|
Klf15
|
Kruppel-like factor 15
|
|
chr11_+_42233258
|
2.323
|
NM_008070
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr4_+_46052090
|
2.287
|
NM_021883
|
Tmod1
|
tropomodulin 1
|
|
chr9_+_111214242
|
2.274
|
NM_001164659
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr13_-_52626065
|
2.272
|
NM_001024474
|
Diras2
|
DIRAS family, GTP-binding RAS-like 2
|
|
chr11_+_105451296
|
2.265
|
NM_181071
|
Tanc2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2
|
|
chr2_-_118855090
|
2.238
|
NM_199310
|
Ccdc32
|
coiled-coil domain containing 32
|
|
chr18_+_37133455
|
2.238
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chrX_+_71002395
|
2.238
|
NM_019587
|
Plxnb3
|
plexin B3
|
|
chr8_-_42219037
|
2.193
|
NM_001005863
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr11_-_105805442
|
2.188
|
NM_007805
|
Cyb561
|
cytochrome b-561
|
|
chr18_+_37303622
|
2.183
|
NM_001003672
|
Pcdhac2
|
protocadherin alpha subfamily C, 2
|
|
chr2_+_158492468
|
2.174
|
NM_153089
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr3_+_31048841
|
2.104
|
NM_008770
|
Cldn11
|
claudin 11
|
|
chr2_+_35413977
|
2.091
|
NM_001114125
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr9_-_95307645
|
2.061
|
NM_018763
|
Chst2
|
carbohydrate sulfotransferase 2
|
|
chr7_+_53651811
|
2.038
|
NM_008421
NM_001112739
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1
|
|
chr1_+_92100572
|
2.027
|
NM_007722
|
Cxcr7
|
chemokine (C-X-C motif) receptor 7
|
|
chr2_-_121097017
|
2.008
|
NM_013735
|
Trp53bp1
|
transformation related protein 53 binding protein 1
|
|
chr8_+_4493404
|
1.987
|
NM_026058
|
Lass4
|
LAG1 homolog, ceramide synthase 4
|
|
chr14_-_66572577
|
1.981
|
NM_172604
|
Scara3
|
scavenger receptor class A, member 3
|
|
chr4_+_101223198
|
1.976
|
NM_001164583
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr8_+_96561097
|
1.973
|
NM_001093757
NM_177767
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1
|
|
chr2_+_181449669
|
1.955
|
NM_011012
|
Oprl1
|
opioid receptor-like 1
|
|
chr1_+_179375951
|
1.951
|
NM_013915
|
Zfp238
|
zinc finger protein 238
|
|
chr7_-_28338127
|
1.951
|
NM_011116
|
Pld3
|
phospholipase D family, member 3
|
|
chr2_-_118529608
|
1.940
|
NM_001081971
|
Gm1337
|
predicted gene 1337
|
|
chr2_+_156246644
|
1.938
|
NM_001003815
NM_013510
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr6_+_135148004
|
1.925
|
NM_028982
|
8430419L09Rik
|
RIKEN cDNA 8430419L09 gene
|
|
chr6_-_113145321
|
1.871
|
NM_177763
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4
|
|
chr13_-_92901406
|
1.829
|
NM_009027
|
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2
|
|
chr6_-_145024596
|
1.826
|
NM_007532
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr9_+_103014374
|
1.791
|
NM_173781
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr16_+_35022488
|
1.780
|
NM_023587
|
Ptplb
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member b
|
|
chr1_+_60466462
|
1.763
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr14_-_56143027
|
1.746
|
NM_008736
|
Nrl
|
neural retina leucine zipper gene
|
|
chr18_+_61085283
|
1.733
|
NM_177407
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr6_-_144997311
|
1.720
|
NM_001024468
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr9_+_59598963
|
1.710
|
NM_173018
|
Myo9a
|
myosin IXa
|
|
chr1_-_134439500
|
1.706
|
NM_177129
|
Cntn2
|
contactin 2
|
|
chrX_-_147531030
|
1.701
|
NM_001164578
NM_175146
|
Tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chrX_+_160346948
|
1.697
|
NM_026887
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit
|
|
chr10_+_18189773
|
1.678
|
NM_001163592
|
Nhsl1
|
NHS-like 1
|
|
chr2_+_48805027
|
1.678
|
NM_029924
|
Mbd5
|
methyl-CpG binding domain protein 5
|
|
chr8_-_85966482
|
1.649
|
NM_019708
|
Scoc
|
short coiled-coil protein
|
|
chr6_-_87285690
|
1.639
|
NM_054041
|
Antxr1
|
anthrax toxin receptor 1
|
|
chr1_+_194014677
|
1.625
|
NM_001038607
NM_010600
|
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr11_-_116060358
|
1.562
|
NM_015729
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl
|
|
chr4_+_151712739
|
1.558
|
NM_001081376
NM_029216
|
Chd5
|
chromodomain helicase DNA binding protein 5
|
|
chr6_+_124948454
|
1.558
|
NM_001145927
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr11_-_95936914
|
1.534
|
NM_001161419
NM_007506
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9)
|
|
chr1_+_72330942
|
1.531
|
NM_175564
|
Tmem169
|
transmembrane protein 169
|
|
chr7_-_147340532
|
1.525
|
NM_183147
|
Sprn
|
shadow of prion protein
|
|
chr2_+_28303460
|
1.517
|
NM_001005420
|
Gm347
|
predicted gene 347
|
|
chr2_-_79296732
|
1.476
|
NM_010894
|
Neurod1
|
neurogenic differentiation 1
|
|
chr14_-_56143799
|
1.476
|
NM_001136074
|
Nrl
|
neural retina leucine zipper gene
|
|
chr13_+_93381086
|
1.465
|
NM_172588
|
Serinc5
|
serine incorporator 5
|
|
chrX_-_133403050
|
1.465
|
NM_176971
|
Rab9b
|
RAB9B, member RAS oncogene family
|
|
chr17_-_24342446
|
1.446
|
NM_001163847
NM_001163848
NM_001163849
NM_001163850
NM_001163851
NM_173186
|
Ntn3
Tbc1d24
|
netrin 3
TBC1 domain family, member 24
|
|
chr6_+_124948627
|
1.445
|
NM_001145926
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr4_+_101180580
|
1.443
|
NM_001164584
NM_001164585
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr13_+_111185251
|
1.433
|
NM_152804
|
Plk2
|
polo-like kinase 2 (Drosophila)
|
|
chr15_+_99501148
|
1.428
|
NM_009597
|
Accn2
|
amiloride-sensitive cation channel 2, neuronal
|
|
chr18_+_86882439
|
1.390
|
NM_172633
|
Cbln2
|
cerebellin 2 precursor protein
|
|
chr7_+_28474358
|
1.383
|
NM_001112731
|
C030039L03Rik
|
RIKEN cDNA C030039L03 gene
|
|
chr7_-_149558139
|
1.381
|
NM_177265
|
6330512M04Rik
|
RIKEN cDNA 6330512M04 gene
|
|
chr8_+_91220921
|
1.373
|
NM_001128170
NM_001128171
NM_173369
NM_001128169
|
Cyld
|
cylindromatosis (turban tumor syndrome)
|
|
chr6_-_121423682
|
1.366
|
NM_001033354
|
Iqsec3
|
IQ motif and Sec7 domain 3
|
|
chr16_-_38713106
|
1.345
|
NM_020260
|
Arhgap31
|
Rho GTPase activating protein 31
|
|
chr2_+_156301543
|
1.336
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr9_+_44925922
|
1.310
|
NM_001014761
|
Scn2b
|
sodium channel, voltage-gated, type II, beta
|
|
chr6_-_137118238
|
1.307
|
NM_001164212
NM_001164214
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor
|
|
chr1_+_94728217
|
1.298
|
NM_016696
|
Gpc1
|
glypican 1
|
|
chr12_+_112127468
|
1.294
|
NM_001081057
|
Tecpr2
|
tectonin beta-propeller repeat containing 2
|
|
chr12_-_88607158
|
1.283
|
NM_001142580
NM_001142581
|
Vipar
|
VPS33B interacting protein, apical-basolateral polarity regulator
|
|
chr3_+_33699111
|
1.273
|
NM_025978
NM_027619
|
Ttc14
|
tetratricopeptide repeat domain 14
|
|
chr8_-_70498472
|
1.258
|
NM_177698
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr17_-_81127339
|
1.251
|
NM_001081357
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3
|
|
chr1_-_38878059
|
1.250
|
NM_001029878
|
Lonrf2
|
LON peptidase N-terminal domain and ring finger 2
|
|
chr15_-_79635138
|
1.244
|
NM_030689
|
Nptxr
|
neuronal pentraxin receptor
|
|
chr17_-_24336901
|
1.243
|
NM_001163852
NM_001163853
|
Tbc1d24
|
TBC1 domain family, member 24
|
|
chr12_+_16901671
|
1.232
|
NM_009072
|
Rock2
|
Rho-associated coiled-coil containing protein kinase 2
|
|
chr4_+_43455122
|
1.226
|
NM_011571
|
Tesk1
|
testis specific protein kinase 1
|
|
chr11_-_74739244
|
1.224
|
NM_001163311
NM_013761
|
Srr
|
serine racemase
|
|
chr2_-_5816381
|
1.224
|
NM_021305
|
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae)
|