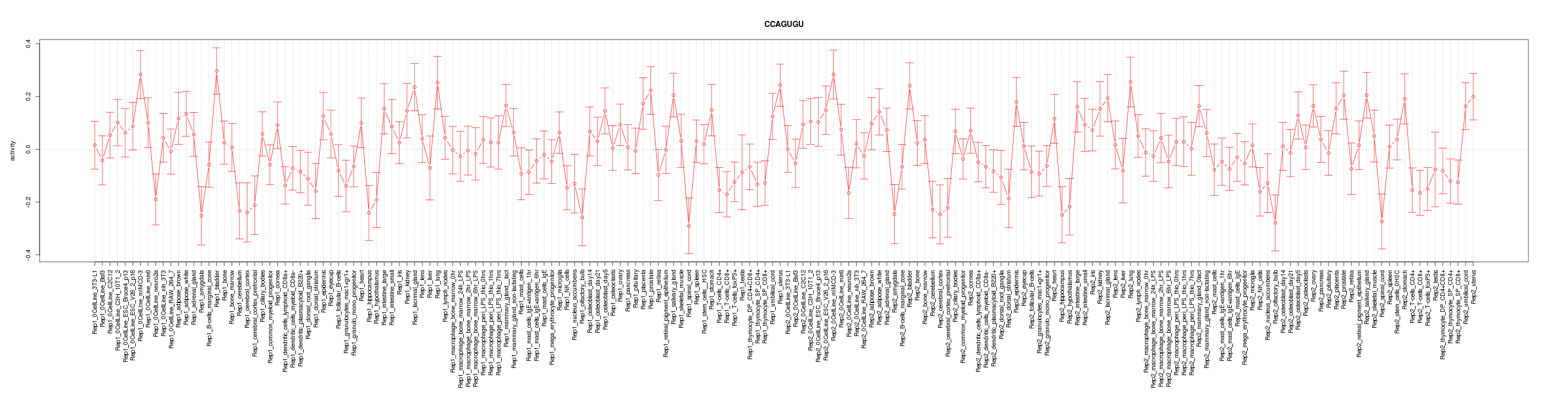
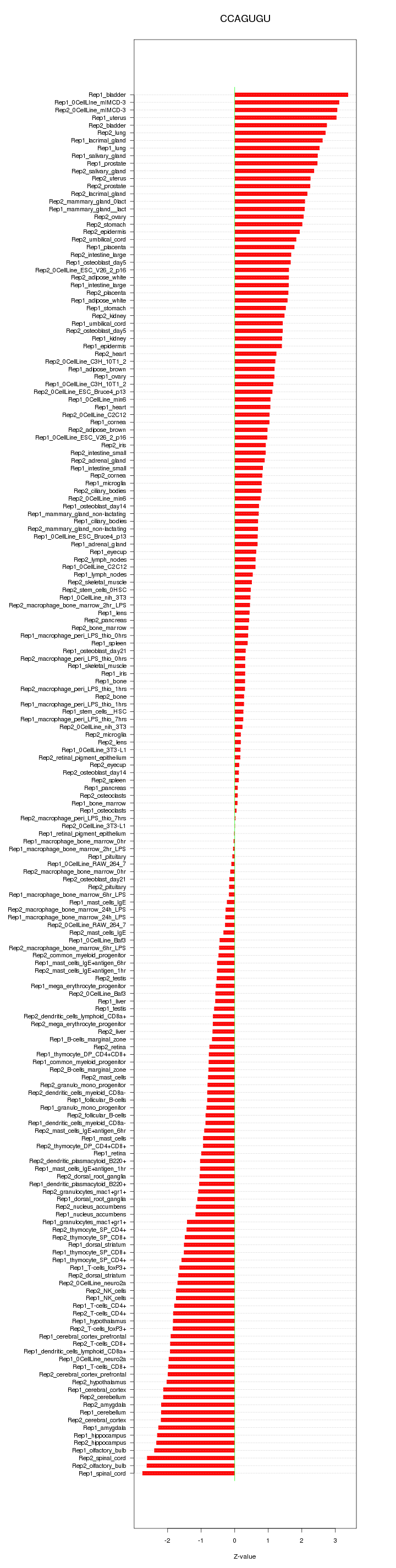
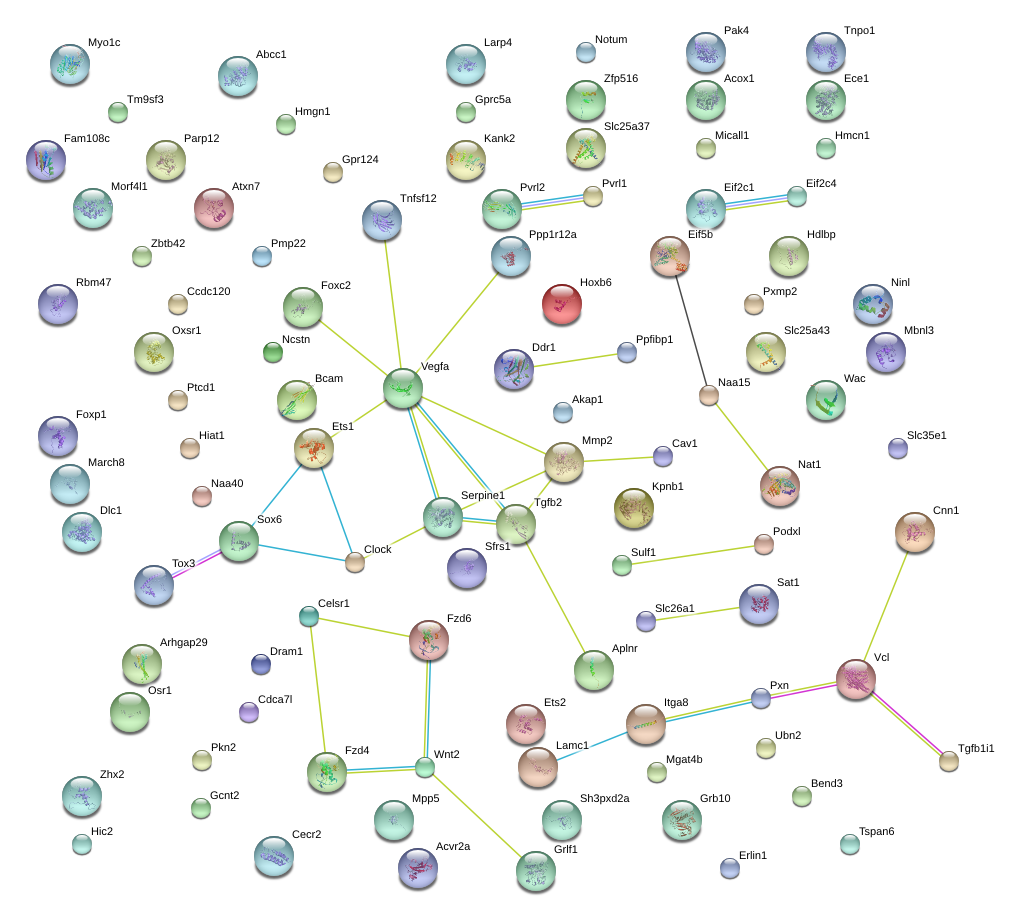
|
chr9_+_21903696
|
32.484
|
NM_009922
|
Cnn1
|
calponin 1
|
|
chr6_+_17256369
|
28.890
|
NM_007616
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr11_+_75465651
|
25.968
|
NM_001080774
|
Myo1c
|
myosin IC
|
|
chr11_+_75465000
|
25.456
|
NM_008659
|
Myo1c
|
myosin IC
|
|
chr11_+_75469516
|
22.569
|
NM_001080775
|
Myo1c
|
myosin IC
|
|
chr7_-_20355880
|
21.058
|
NM_020486
|
Bcam
|
basal cell adhesion molecule
|
|
chr5_-_66489416
|
19.973
|
NM_139065
|
Rbm47
|
RNA binding motif protein 47
|
|
chr15_+_38837839
|
19.515
|
NM_008056
|
Fzd6
|
frizzled homolog 6 (Drosophila)
|
|
chr15_+_38837818
|
18.819
|
NM_001162494
|
Fzd6
|
frizzled homolog 6 (Drosophila)
|
|
chr5_-_66543123
|
18.070
|
NM_001127382
|
Rbm47
|
RNA binding motif protein 47
|
|
chr8_+_28196312
|
17.596
|
NM_054044
|
Gpr124
|
G protein-coupled receptor 124
|
|
chr12_+_9581218
|
17.095
|
NM_011859
|
Osr1
|
odd-skipped related 1 (Drosophila)
|
|
chr11_-_88725877
|
16.471
|
NM_001042541
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr5_-_66542161
|
15.713
|
NM_178446
|
Rbm47
|
RNA binding motif protein 47
|
|
chr8_+_95351212
|
15.486
|
NM_008610
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr17_-_35840931
|
15.329
|
NM_007584
NM_172962
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr1_-_155179844
|
14.604
|
NM_010683
|
Lamc1
|
laminin, gamma 1
|
|
chr6_+_135015679
|
13.670
|
NM_181444
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr5_-_137548123
|
12.704
|
NM_008871
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chrX_-_130432840
|
12.560
|
NM_019656
|
Tspan6
|
tetraspanin 6
|
|
chr6_-_39068340
|
12.320
|
NM_172893
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr11_-_11937330
|
12.258
|
NM_010345
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr6_+_38383918
|
11.893
|
NM_177185
|
Ubn2
|
ubinuclein 2
|
|
chr5_+_115956682
|
11.808
|
NM_011223
NM_133915
|
Pxn
|
paxillin
|
|
chr3_+_121656136
|
11.740
|
NM_172525
|
Arhgap29
|
Rho GTPase activating protein 29
|
|
chr13_+_40982534
|
11.423
|
NM_008105
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme
|
|
chr11_-_11927973
|
11.110
|
NM_001177629
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr16_+_95923988
|
11.078
|
NM_011809
|
Ets2
|
E26 avian leukemia oncogene 2, 3' domain
|
|
chr11_-_88712812
|
11.007
|
NM_009648
|
Akap1
|
A kinase (PRKA) anchor protein 1
|
|
chr1_+_12682510
|
10.875
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr6_-_31513817
|
10.815
|
NM_013723
|
Podxl
|
podocalyxin-like
|
|
chr6_+_116288075
|
10.813
|
NM_027920
|
March8
|
membrane-associated ring finger (C3HC4) 8
|
|
chr11_+_62944973
|
10.724
|
NM_008885
|
Pmp22
|
peripheral myelin protein 22
|
|
chr1_+_12708582
|
10.472
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr1_-_95375372
|
10.357
|
NM_133808
|
Hdlbp
|
high density lipoprotein (HDL) binding protein
|
|
chr11_-_69499014
|
10.213
|
NM_001159505
NM_023517
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr10_+_43198945
|
10.062
|
NM_199028
|
Bend3
|
BEN domain containing 3
|
|
chr18_+_83084023
|
9.999
|
NM_001177464
|
Zfp516
|
zinc finger protein 516
|
|
chr2_-_12223518
|
9.599
|
NM_001001309
|
Itga8
|
integrin alpha 8
|
|
chr13_+_40955500
|
8.982
|
NM_023887
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme
|
|
chr1_-_188529867
|
8.908
|
NM_009367
|
Tgfb2
|
transforming growth factor, beta 2
|
|
chr12_+_79849918
|
8.742
|
NM_019579
|
Mpp5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr9_-_21602942
|
8.704
|
NM_145611
|
Kank2
|
KN motif and ankyrin repeat domains 2
|
|
chr15_-_85864206
|
8.448
|
NM_009886
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila)
|
|
chr7_-_29383180
|
8.273
|
NM_027470
|
Pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr10_-_87819765
|
8.239
|
NM_027878
|
Dram1
|
DNA-damage regulated autophagy modulator 1
|
|
chr14_+_21748646
|
7.830
|
NM_009502
|
Vcl
|
vinculin
|
|
chr8_-_38001686
|
7.515
|
NM_001194940
|
Dlc1
|
deleted in liver cancer 1
|
|
chr7_+_96552871
|
7.439
|
NM_008055
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr7_-_91300359
|
7.392
|
NM_133722
|
Fam108c
|
family with sequence similarity 108, member C
|
|
chr5_-_76733540
|
7.321
|
NM_007715
|
Clock
|
circadian locomotor output cycles kaput
|
|
chr19_-_47538898
|
7.264
|
NM_001164717
NM_008018
|
Sh3pxd2a
|
SH3 and PX domains 2A
|
|
chr13_-_99696254
|
7.157
|
NM_178716
|
Tnpo1
|
transportin 1
|
|
chr14_-_69903072
|
7.124
|
NM_026331
|
Slc25a37
|
solute carrier family 25, member 37
|
|
chr6_+_146837012
|
7.100
|
NM_001170433
NM_026221
|
Ppfibp1
|
PTPRF interacting protein, binding protein 1 (liprin beta 1)
|
|
chr18_+_83080270
|
7.081
|
NM_183033
|
Zfp516
|
zinc finger protein 516
|
|
chr7_-_20334872
|
6.826
|
NM_001159724
NM_008990
|
Pvrl2
|
poliovirus receptor-related 2
|
|
chr15_+_78939343
|
6.747
|
NM_177461
|
Micall1
|
microtubule associated monoxygenase, calponin and LIM domain containing -like 1
|
|
chr7_-_123182257
|
6.729
|
NM_001025559
|
Sox6
|
SRY-box containing gene 6
|
|
chr2_+_48669628
|
6.535
|
NM_007396
|
Acvr2a
|
activin receptor IIA
|
|
chr7_+_135390325
|
6.512
|
NM_009365
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr11_+_96160484
|
6.394
|
NM_008269
|
Hoxb6
|
homeobox B6
|
|
chr17_-_35837432
|
6.379
|
NM_001198833
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr10_+_107599446
|
6.337
|
NM_027892
|
Ppp1r12a
|
protein phosphatase 1, regulatory (inhibitor) subunit 12A
|
|
chr13_+_41012969
|
6.335
|
NM_133219
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme
|
|
chr11_+_50038815
|
6.320
|
NM_145926
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B
|
|
chr13_-_99660903
|
6.292
|
NM_001048267
|
Tnpo1
|
transportin 1
|
|
chr8_-_37796104
|
6.277
|
NM_001194941
|
Dlc1
|
deleted in liver cancer 1
|
|
chr17_-_46169322
|
6.194
|
NM_001025250
NM_001025257
NM_009505
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr11_-_69509498
|
6.160
|
NM_011614
NM_001034097
NM_001159503
|
Tnfsf12
BC096441
|
tumor necrosis factor (ligand) superfamily, member 12
cDNA sequence BC096441
|
|
chr1_-_174012771
|
6.085
|
NM_021607
|
Ncstn
|
nicastrin
|
|
chr3_-_142544883
|
5.762
|
NM_178654
|
Pkn2
|
protein kinase N2
|
|
chr19_-_44144244
|
5.749
|
NM_001164359
NM_001164360
|
Erlin1
|
ER lipid raft associated 1
|
|
chr6_+_120616386
|
5.699
|
NM_001128151
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 homolog (human)
|
|
chr19_-_41338400
|
5.647
|
NM_133352
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chrX_-_151650871
|
5.575
|
NM_009121
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1
|
|
chr6_-_98978185
|
5.570
|
NM_001197322
|
Foxp1
|
forkhead box P1
|
|
chr1_-_152840432
|
5.479
|
NM_001024720
|
Hmcn1
|
hemicentin 1
|
|
chrX_-_48559008
|
5.263
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr4_-_126145660
|
5.223
|
NM_153403
|
Eif2c1
|
eukaryotic translation initiation factor 2C, 1
|
|
chr4_+_137418151
|
5.211
|
NM_199307
|
Ece1
|
endothelin converting enzyme 1
|
|
chr11_-_97049185
|
5.126
|
NM_008379
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chr8_+_123640069
|
5.110
|
NM_013519
|
Foxc2
|
forkhead box C2
|
|
chr6_-_99385338
|
4.949
|
NM_053202
|
Foxp1
|
forkhead box P1
|
|
chr6_-_99216494
|
4.945
|
NM_001197321
|
Foxp1
|
forkhead box P1
|
|
chr3_-_116415169
|
4.849
|
NM_144902
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3
|
|
chr12_+_113917038
|
4.804
|
NM_001100460
|
Zbtb42
|
zinc finger and BTB domain containing 42
|
|
chr7_-_17200285
|
4.662
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr2_+_84976516
|
4.645
|
NM_011784
|
Aplnr
|
apelin receptor
|
|
chr11_-_120522148
|
4.460
|
NM_175263
|
Notum
|
notum pectinacetylesterase homolog (Drosophila)
|
|
chr9_+_43552516
|
4.397
|
NM_021424
|
Pvrl1
|
poliovirus receptor-related 1
|
|
chr3_+_51219882
|
4.362
|
NM_053089
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit
|
|
chr12_+_119082333
|
4.253
|
NM_146040
|
Cdca7l
|
cell division cycle associated 7 like
|
|
chr8_-_92872139
|
4.213
|
NM_172913
|
Tox3
|
TOX high mobility group box family member 3
|
|
chr16_+_17233664
|
4.151
|
NM_178922
|
Hic2
|
hypermethylated in cancer 2
|
|
chr17_-_46162111
|
4.131
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr14_+_14845004
|
4.058
|
NM_139227
|
Atxn7
|
ataxin 7
|
|
chrX_-_7318412
|
4.046
|
NM_207202
|
Ccdc120
|
coiled-coil domain containing 120
|
|
chr1_+_38054854
|
4.000
|
NM_198303
|
Eif5b
|
eukaryotic translation initiation factor 5B
|
|
chr16_+_14361637
|
3.964
|
NM_008576
|
Abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr16_-_96349292
|
3.890
|
NM_008251
|
Hmgn1
|
high mobility group nucleosomal binding domain 1
|
|
chr6_-_17980373
|
3.868
|
NM_023653
|
Wnt2
|
wingless-related MMTV integration site 2
|
|
chr7_-_123174560
|
3.867
|
NM_001025560
|
Sox6
|
SRY-box containing gene 6
|
|
chr19_-_7315495
|
3.807
|
NM_027643
|
Naa40
|
N(alpha)-acetyltransferase 40, NatD catalytic subunit, homolog (S. cerevisiae)
|
|
chr11_-_116060358
|
3.727
|
NM_015729
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl
|
|
chr15_+_99800504
|
3.663
|
NM_001080948
|
Larp4
|
La ribonucleoprotein domain family, member 4
|
|
chr8_-_75016484
|
3.560
|
NM_177766
|
Slc35e1
|
solute carrier family 35, member E1
|
|
chr2_-_150835067
|
3.547
|
NM_207204
|
Ninl
|
ninein-like
|
|
chr11_+_87861165
|
3.525
|
NM_001078167
NM_173374
|
Srsf1
|
serine/arginine-rich splicing factor 1
|
|
chr19_+_5689130
|
3.524
|
NM_022012
|
Map3k11
|
mitogen-activated protein kinase kinase kinase 11
|
|
chr15_+_99803210
|
3.474
|
NM_001024526
|
Larp4
|
La ribonucleoprotein domain family, member 4
|
|
chr11_+_69794546
|
3.356
|
NM_026017
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1
|
|
chr9_-_21397188
|
3.344
|
NM_138303
|
Yipf2
|
Yip1 domain family, member 2
|
|
chrX_-_101176600
|
3.308
|
NM_011276
|
Rlim
|
ring finger protein, LIM domain interacting
|
|
chr8_-_37676995
|
3.290
|
NM_015802
|
Dlc1
|
deleted in liver cancer 1
|
|
chr7_-_89137131
|
3.237
|
NM_007562
|
Bnc1
|
basonuclin 1
|
|
chr9_-_20619426
|
3.217
|
NM_016919
|
Col5a3
|
collagen, type V, alpha 3
|
|
chrX_+_12858142
|
3.172
|
NM_010028
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr13_+_47138896
|
3.138
|
NM_172262
|
Kdm1b
|
lysine (K)-specific demethylase 1B
|
|
chr11_-_69761489
|
3.121
|
NM_009204
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr7_-_123138599
|
3.083
|
NM_011445
|
Sox6
|
SRY-box containing gene 6
|
|
chr17_-_35838974
|
3.032
|
NM_001198831
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr4_-_154010868
|
3.015
|
NM_001177995
NM_027504
|
Prdm16
|
PR domain containing 16
|
|
chr12_+_80130149
|
3.012
|
NM_181073
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1
|
|
chrX_-_103207106
|
2.889
|
NM_001190409
NM_025952
|
Magt1
|
magnesium transporter 1
|
|
chr8_-_108351060
|
2.876
|
NM_145824
|
Ranbp10
|
RAN binding protein 10
|
|
chr19_+_5038825
|
2.867
|
NM_175383
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr4_-_138648762
|
2.865
|
NM_008675
|
Nbl1
|
neuroblastoma, suppression of tumorigenicity 1
|
|
chr6_-_115944822
|
2.843
|
NM_026376
|
Plxnd1
|
plexin D1
|
|
chr17_+_14416508
|
2.803
|
NM_022315
|
Smoc2
|
SPARC related modular calcium binding 2
|
|
chr4_+_13678443
|
2.764
|
NM_001111027
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related)
|
|
chr13_+_112476313
|
2.535
|
NM_172593
|
Mier3
|
mesoderm induction early response 1, family member 3
|
|
chr15_-_85412250
|
2.526
|
NM_009528
|
Wnt7b
|
wingless-related MMTV integration site 7B
|
|
chr19_-_60656627
|
2.520
|
NM_001172096
NM_001172097
NM_030197
|
2700078E11Rik
|
RIKEN cDNA 2700078E11 gene
|
|
chr11_-_120434204
|
2.519
|
NM_011032
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr15_-_85408499
|
2.480
|
NM_001163634
|
Wnt7b
|
wingless-related MMTV integration site 7B
|
|
chr4_-_44181148
|
2.422
|
NM_001038993
|
Rnf38
|
ring finger protein 38
|
|
chr16_+_4594703
|
2.361
|
NM_031184
|
Glis2
|
GLIS family zinc finger 2
|
|
chr2_-_155555105
|
2.340
|
NM_145537
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2
|
|
chr13_-_53076251
|
2.326
|
NM_017373
|
Nfil3
|
nuclear factor, interleukin 3, regulated
|
|
chr14_+_118536326
|
2.326
|
NM_021434
|
Gpr180
|
G protein-coupled receptor 180
|
|
chr11_+_77499638
|
2.321
|
NM_001024205
|
Nufip2
|
nuclear fragile X mental retardation protein interacting protein 2
|
|
chr10_+_51953390
|
2.308
|
NM_025705
|
Dcbld1
|
discoidin, CUB and LCCL domain containing 1
|
|
chr9_-_114473486
|
2.303
|
NM_001042503
|
Trim71
|
tripartite motif-containing 71
|
|
chr17_+_13897509
|
2.240
|
NM_010806
|
Mllt4
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 4
|
|
chr9_-_71619329
|
2.212
|
NM_026599
|
Cgnl1
|
cingulin-like 1
|
|
chr4_-_132066370
|
2.162
|
NM_144907
|
Sesn2
|
sestrin 2
|
|
chr2_+_3630729
|
2.132
|
NM_025626
|
Fam107b
|
family with sequence similarity 107, member B
|
|
chr18_-_10181789
|
2.107
|
NM_009071
|
Rock1
|
Rho-associated coiled-coil containing protein kinase 1
|
|
chr6_+_122258946
|
2.091
|
NM_010749
|
M6pr
|
mannose-6-phosphate receptor, cation dependent
|
|
chr3_-_9610062
|
2.076
|
NM_133218
|
Zfp704
|
zinc finger protein 704
|
|
chrX_+_98469429
|
2.053
|
NM_021521
|
Med12
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast)
|
|
chr2_-_103637096
|
2.040
|
NM_001111289
NM_001111290
NM_001111292
|
Caprin1
|
cell cycle associated protein 1
|
|
chr16_+_33380860
|
2.030
|
NM_011749
|
Zfp148
|
zinc finger protein 148
|
|
chr2_-_93174606
|
2.013
|
NM_183180
|
Tspan18
|
tetraspanin 18
|
|
chr10_-_120913138
|
2.004
|
NM_138956
|
Rassf3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr4_-_109149038
|
1.984
|
NM_013876
|
Rnf11
|
ring finger protein 11
|
|
chr19_-_57083064
|
1.936
|
NM_001177796
NM_001177797
NM_146102
|
Afap1l2
|
actin filament associated protein 1-like 2
|
|
chr17_+_78599556
|
1.921
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr16_-_31274947
|
1.862
|
NM_025800
|
Ppp1r2
|
protein phosphatase 1, regulatory (inhibitor) subunit 2
|
|
chr19_-_41876509
|
1.849
|
NM_001163495
NM_027667
|
Arhgap19
|
Rho GTPase activating protein 19
|
|
chr14_+_31832323
|
1.833
|
NM_001081251
|
Pbrm1
|
polybromo 1
|
|
chr6_-_38826156
|
1.787
|
NM_010433
|
Hipk2
|
homeodomain interacting protein kinase 2
|
|
chr7_+_80520360
|
1.772
|
NM_177740
|
Rgma
|
RGM domain family, member A
|
|
chr5_+_139676589
|
1.750
|
NM_024451
|
Sun1
|
Sad1 and UNC84 domain containing 1
|
|
chr1_-_39535525
|
1.737
|
NM_018775
|
Tbc1d8
|
TBC1 domain family, member 8
|
|
chr12_-_93024875
|
1.721
|
NM_175367
|
Ston2
|
stonin 2
|
|
chr10_-_62801726
|
1.695
|
NM_001159590
NM_019812
NM_001159589
|
Sirt1
|
sirtuin 1 (silent mating type information regulation 2, homolog) 1 (S. cerevisiae)
|
|
chr5_+_148242018
|
1.680
|
NM_028291
|
Pan3
|
PAN3 polyA specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr3_-_65762057
|
1.626
|
NM_019937
|
Ccnl1
|
cyclin L1
|
|
chr9_+_21420612
|
1.557
|
NM_001174078
NM_001174079
NM_011417
|
Smarca4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4
|
|
chr17_-_66450782
|
1.546
|
NM_028388
|
Ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2
|
|
chr8_+_36650839
|
1.524
|
NM_001081279
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr4_-_44180323
|
1.395
|
NM_175201
|
Rnf38
|
ring finger protein 38
|
|
chr11_-_59104252
|
1.385
|
NM_009522
|
Wnt3a
|
wingless-related MMTV integration site 3A
|
|
chr2_-_160192799
|
1.324
|
NM_010658
|
Mafb
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein B (avian)
|
|
chr10_+_127176590
|
1.317
|
NM_198035
|
Zbtb39
|
zinc finger and BTB domain containing 39
|
|
chr11_-_45979836
|
1.314
|
NM_172524
|
Nipal4
|
NIPA-like domain containing 4
|
|
chr15_-_77672472
|
1.310
|
NM_022410
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle
|
|
chr4_-_53172748
|
1.305
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr2_-_140497204
|
1.301
|
NM_001172160
NM_178382
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_+_133985717
|
1.227
|
NM_007961
|
Etv6
|
ets variant gene 6 (TEL oncogene)
|
|
chrX_+_130442964
|
1.165
|
NM_001083895
NM_026838
|
Srpx2
|
sushi-repeat-containing protein, X-linked 2
|
|
chr16_+_44173356
|
1.119
|
NM_001029889
|
Gm608
|
predicted gene 608
|
|
chr18_-_63851841
|
1.115
|
NM_016792
|
Txnl1
|
thioredoxin-like 1
|
|
chr11_-_104303565
|
1.089
|
NM_001081045
|
1700081L11Rik
|
RIKEN cDNA 1700081L11 gene
|
|
chr4_-_43512476
|
1.079
|
NM_001013377
|
E130306D19Rik
|
RIKEN cDNA E130306D19 gene
|
|
chr13_-_46060325
|
1.073
|
NM_009124
|
Atxn1
|
ataxin 1
|
|
chr2_+_138082301
|
1.066
|
NM_001025431
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr2_+_34627607
|
1.033
|
NM_001163434
NM_022310
|
Hspa5
|
heat shock protein 5
|
|
chr4_-_10934608
|
1.008
|
NM_175175
|
Plekhf2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2
|
|
chr7_-_105326687
|
0.929
|
NM_007602
|
Capn5
|
calpain 5
|
|
chr3_+_135148555
|
0.910
|
NM_027288
|
Manba
|
mannosidase, beta A, lysosomal
|
|
chr4_+_115823089
|
0.895
|
NM_026651
NM_029786
|
Pomgnt1
|
protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase
|
|
chr8_+_81033220
|
0.889
|
NM_029736
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7
|
|
chr4_+_55362895
|
0.882
|
NM_009011
|
Rad23b
|
RAD23b homolog (S. cerevisiae)
|
|
chr15_-_100253413
|
0.879
|
NM_001146161
NM_008732
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr19_+_54018795
|
0.864
|
NM_001168505
|
Shoc2
|
soc-2 (suppressor of clear) homolog (C. elegans)
|
|
chr7_-_35070627
|
0.854
|
NM_172741
|
4931406P16Rik
|
RIKEN cDNA 4931406P16 gene
|
|
chr15_+_102038697
|
0.821
|
NM_153194
|
Zfp740
|
zinc finger protein 740
|