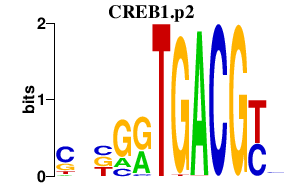
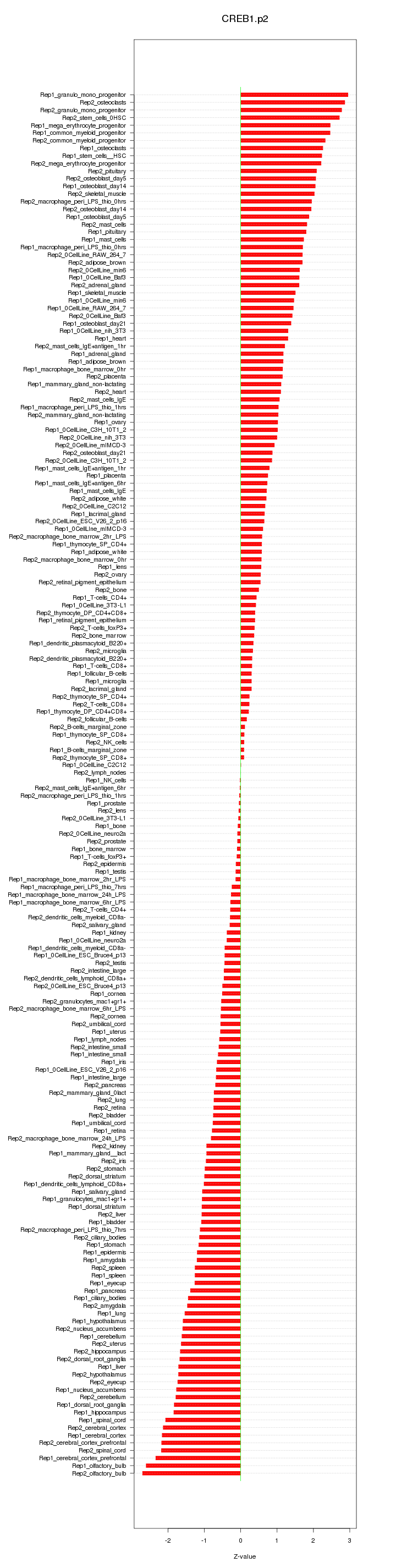
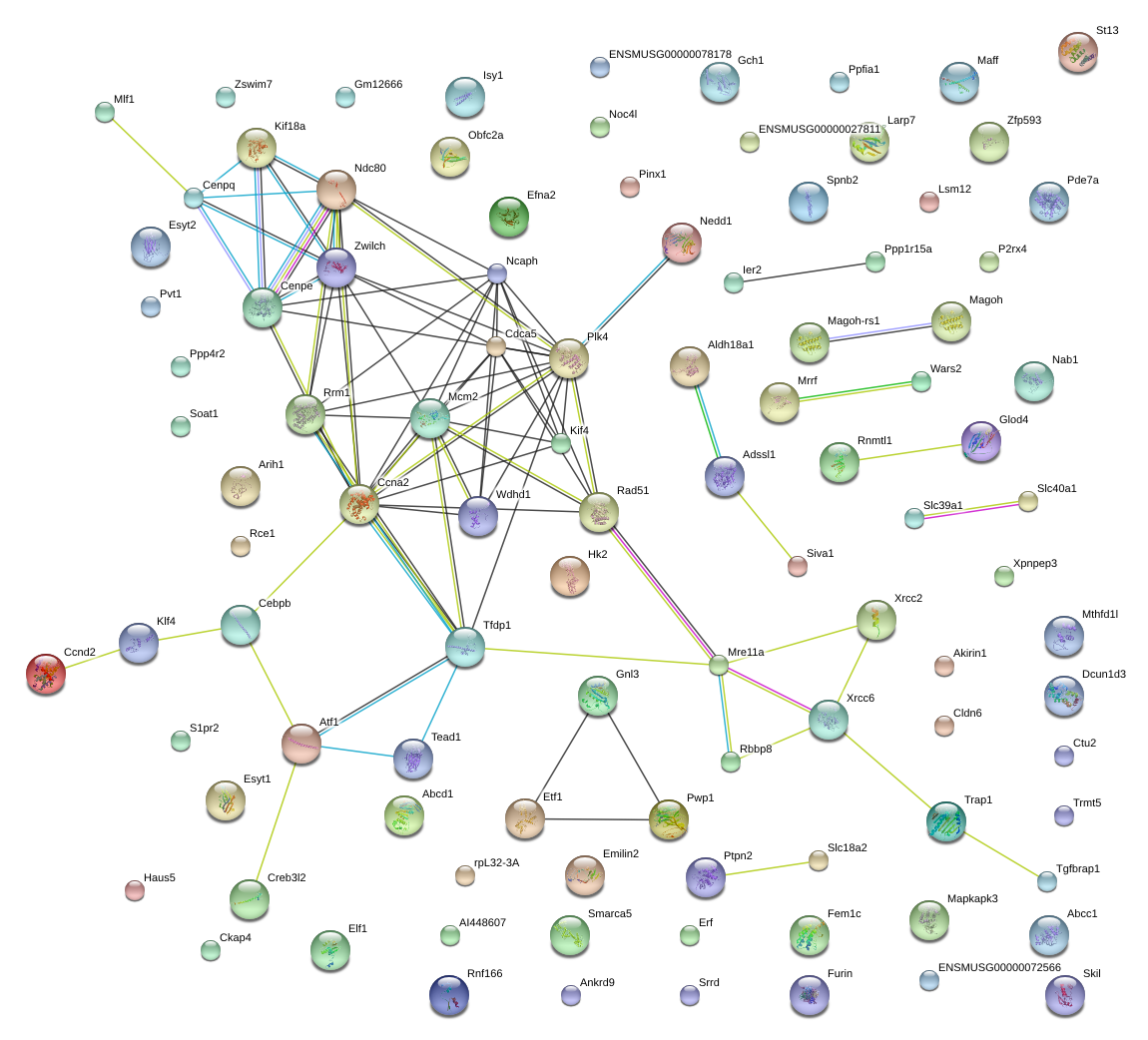
|
chr8_+_13338741
|
14.173
|
|
Tfdp1
|
transcription factor Dp 1
|
|
chr18_+_11815869
|
13.941
|
|
Rbbp8
|
retinoblastoma binding protein 8
|
|
chr3_-_19211000
|
13.566
|
NM_001122759
|
Pde7a
|
phosphodiesterase 7A
|
|
chr6_-_88848650
|
13.365
|
NM_008564
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae)
|
|
chr7_-_31449991
|
12.848
|
NM_027999
|
Haus5
|
HAUS augmin-like complex, subunit 5
|
|
chr2_+_35992148
|
11.968
|
|
Mrrf
|
mitochondrial ribosome recycling factor
|
|
chr19_-_4625574
|
11.356
|
NM_023131
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr19_-_4625276
|
10.949
|
|
Rce1
|
RCE1 homolog, prenyl protein peptidase (S. cerevisiae)
|
|
chr3_-_36470904
|
10.712
|
NM_009828
|
Ccna2
|
cyclin A2
|
|
chr11_-_76057153
|
10.355
|
|
Glod4
|
glyoxalase domain containing 4
|
|
chr3_-_36470845
|
10.174
|
|
Ccna2
|
cyclin A2
|
|
chr7_-_31449951
|
9.936
|
|
Haus5
|
HAUS augmin-like complex, subunit 5
|
|
chr4_-_133801438
|
9.781
|
NM_024215
|
Zfp593
|
zinc finger protein 593
|
|
chr12_+_113883038
|
9.407
|
NM_001161737
NM_013929
|
Siva1
|
SIVA1, apoptosis-inducing factor
|
|
chr3_+_134875526
|
9.305
|
NM_173762
|
Cenpe
|
centromere protein E
|
|
chr3_-_36470882
|
9.104
|
|
Ccna2
|
cyclin A2
|
|
chr9_-_64020685
|
8.860
|
NM_026507
|
Zwilch
|
Zwilch, kinetochore associated, homolog (Drosophila)
|
|
chr17_-_71660199
|
8.766
|
NM_145158
|
Emilin2
|
elastin microfibril interfacer 2
|
|
chr15_+_100058249
|
8.750
|
NM_007497
|
Atf1
|
activating transcription factor 1
|
|
chr11_-_76057188
|
8.630
|
NM_026029
|
Glod4
|
glyoxalase domain containing 4
|
|
chr3_-_127256215
|
8.611
|
NM_138593
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr6_-_82724366
|
8.295
|
NM_013820
|
Hk2
|
hexokinase 2
|
|
chr19_+_59335367
|
8.290
|
NM_172523
|
Slc18a2
|
solute carrier family 18 (vesicular monoamine), member 2
|
|
chr2_+_118938549
|
8.271
|
NM_011234
|
Rad51
|
RAD51 homolog (S. cerevisiae)
|
|
chr2_-_126959605
|
8.207
|
NM_144818
|
Ncaph
|
non-SMC condensin I complex, subunit H
|
|
chr12_+_117519658
|
7.985
|
NM_028731
|
Esyt2
|
extended synaptotagmin-like protein 2
|
|
chr3_+_30993952
|
7.876
|
NM_001039090
NM_011386
|
Skil
|
SKI-like
|
|
chr14_+_79880952
|
7.811
|
NM_007920
|
Elf1
|
E74-like factor 1
|
|
chr11_-_62094853
|
7.736
|
NM_027198
|
Zswim7
|
zinc finger, SWIM-type containing 7
|
|
chr19_+_6085096
|
7.474
|
NM_026410
|
Cdca5
|
cell division cycle associated 5
|
|
chr17_-_71876127
|
7.455
|
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr7_+_109590208
|
7.453
|
NM_009103
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr15_+_81846798
|
7.451
|
NM_010247
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr15_+_79177959
|
7.449
|
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr6_-_131243197
|
7.400
|
NM_025564
|
Magohb
|
mago-nashi homolog B (Drosophila)
|
|
chr17_-_71876150
|
7.295
|
NM_023294
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr14_-_47896531
|
7.270
|
NM_172598
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr15_-_81229961
|
7.243
|
|
St13
|
suppression of tumorigenicity 13
|
|
chr2_+_109120894
|
7.221
|
NM_139303
|
Kif18a
|
kinesin family member 18A
|
|
chr11_+_76057230
|
7.203
|
NM_183263
|
Rnmtl1
|
RNA methyltransferase like 1
|
|
chr10_-_6373388
|
7.043
|
NM_001170785
NM_001170786
NM_172308
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr15_+_100058310
|
6.947
|
|
Atf1
|
activating transcription factor 1
|
|
chr15_+_79178063
|
6.898
|
NM_010755
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian)
|
|
chr8_+_125000042
|
6.819
|
NM_153775
|
Ctu2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe)
|
|
chr2_+_167514414
|
6.738
|
NM_009883
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr16_-_4077714
|
6.673
|
NM_026508
|
Trap1
|
TNF receptor-associated protein 1
|
|
chr3_+_40603872
|
6.558
|
NM_011495
|
Plk4
|
polo-like kinase 4 (Drosophila)
|
|
chr6_-_127100954
|
6.541
|
NM_009829
|
Ccnd2
|
cyclin D2
|
|
chr7_-_52781502
|
6.449
|
NM_008654
|
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A
|
|
chr10_-_6373309
|
6.402
|
|
Mthfd1l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like
|
|
chr9_-_107192203
|
6.355
|
NM_178907
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr14_-_31832165
|
6.354
|
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr3_+_67178037
|
6.319
|
|
Mlf1
|
myeloid leukemia factor 1
|
|
chr5_-_111082387
|
6.310
|
NM_153570
|
Noc4l
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr11_-_102046532
|
6.284
|
NM_172947
|
Lsm12
|
LSM12 homolog (S. cerevisiae)
|
|
chr8_-_87186691
|
6.279
|
NM_010499
|
Ier2
|
immediate early response 2
|
|
chr17_-_41071432
|
6.204
|
NM_031863
|
Cenpq
|
centromere protein Q
|
|
chr8_-_83263323
|
6.131
|
NM_053124
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr17_+_23816331
|
6.130
|
NM_018777
|
Cldn6
|
claudin 6
|
|
chr7_+_119822831
|
6.061
|
NM_001166584
NM_009346
|
Tead1
|
TEA domain family member 1
|
|
chr9_-_20781206
|
6.041
|
NM_010333
|
S1pr2
|
sphingosine-1-phosphate receptor 2
|
|
chr9_+_14589080
|
6.023
|
|
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae)
|
|
chr9_-_59334393
|
5.960
|
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila)
|
|
chr14_+_64479167
|
5.945
|
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1
|
|
chr14_-_31832274
|
5.933
|
NM_153547
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar)
|
|
chr9_-_20781133
|
5.921
|
|
S1pr2
|
sphingosine-1-phosphate receptor 2
|
|
chr3_+_67178016
|
5.902
|
NM_001039543
NM_010801
|
Mlf1
|
myeloid leukemia factor 1
|
|
chr3_+_79433000
|
5.894
|
NM_029482
|
4930579G24Rik
|
RIKEN cDNA 4930579G24 gene
|
|
chr10_-_92185071
|
5.841
|
NM_008682
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated gene 1
|
|
chr9_+_14589122
|
5.829
|
NM_018736
|
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae)
|
|
chr3_+_90052083
|
5.804
|
NM_013901
|
Slc39a1
|
solute carrier family 39 (zinc transporter), member 1
|
|
chr9_-_59334089
|
5.780
|
NM_019927
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila)
|
|
chr3_+_98944990
|
5.724
|
NM_027462
|
Wars2
|
tryptophanyl tRNA synthetase 2 (mitochondrial)
|
|
chr8_-_124999921
|
5.720
|
NM_001033142
|
Rnf166
|
ring finger protein 166
|
|
chr4_-_55545337
|
5.682
|
NM_010637
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr10_+_85334563
|
5.678
|
NM_133993
|
Pwp1
|
PWP1 homolog (S. cerevisiae)
|
|
chr2_+_167514460
|
5.594
|
|
Cebpb
|
CCAAT/enhancer binding protein (C/EBP), beta
|
|
chr6_-_87788752
|
5.555
|
NM_133934
|
Isy1
|
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr1_-_158404299
|
5.514
|
NM_009230
|
Soat1
|
sterol O-acyltransferase 1
|
|
chr9_-_107191674
|
5.488
|
|
Mapkapk3
|
mitogen-activated protein kinase-activated protein kinase 3
|
|
chr4_-_123427465
|
5.481
|
NM_023423
|
Akirin1
|
akirin 1
|
|
chr5_-_112772024
|
5.469
|
NM_027323
|
Srrd
|
SRR1 domain containing
|
|
chr14_-_47809063
|
5.442
|
NM_008102
|
Gch1
|
GTP cyclohydrolase 1
|
|
chr12_+_113858217
|
5.410
|
NM_007421
|
Adssl1
|
adenylosuccinate synthetase like 1
|
|
chr15_+_81230560
|
5.247
|
NM_177310
|
Xpnpep3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative
|
|
chr3_-_127256181
|
5.214
|
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr15_+_61869529
|
5.208
|
|
Pvt1
|
plasmacytoma variant translocation 1
|
|
chr18_+_11816327
|
5.197
|
NM_001081223
|
Rbbp8
|
retinoblastoma binding protein 8
|
|
chr7_-_151739579
|
5.190
|
NM_001033319
NM_001195086
|
Ppfia1
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1
|
|
chr11_+_69794546
|
5.160
|
NM_026017
|
Ctdnep1
|
CTD nuclear envelope phosphatase 1
|
|
chr19_-_40662819
|
5.154
|
NM_019698
NM_153554
|
Aldh18a1
|
aldehyde dehydrogenase 18 family, member A1
|
|
chr7_-_87547589
|
5.051
|
NM_011046
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr7_-_26035683
|
5.013
|
NM_010155
|
Erf
|
Ets2 repressor factor
|
|
chr6_-_87788729
|
4.986
|
|
Isy1
|
ISY1 splicing factor homolog (S. cerevisiae)
|
|
chr16_+_14361637
|
4.981
|
NM_008576
|
Abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr19_+_18745215
|
4.966
|
NM_026120
|
2410127L17Rik
|
RIKEN cDNA 2410127L17 gene
|
|
chr8_-_83262881
|
4.948
|
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chrX_+_97821403
|
4.945
|
NM_008446
|
Kif4
|
kinesin family member 4
|
|
chr3_+_127256406
|
4.925
|
NM_197997
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene
|
|
chr5_+_123157541
|
4.876
|
NM_011026
|
P2rx4
|
purinergic receptor P2X, ligand-gated ion channel 4
|
|
chr7_-_127039189
|
4.859
|
NM_001163703
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr18_-_46685512
|
4.839
|
|
Fem1c
|
fem-1 homolog c (C.elegans)
|
|
chr3_-_127256114
|
4.805
|
|
Larp7
|
La ribonucleoprotein domain family, member 7
|
|
chr12_-_112217151
|
4.790
|
NM_175207
|
Ankrd9
|
ankyrin repeat domain 9
|
|
chr1_-_51535232
|
4.782
|
NM_028696
|
Obfc2a
|
oligonucleotide/oligosaccharide-binding fold containing 2A
|
|
chr6_+_100783598
|
4.774
|
NM_182939
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr10_-_83996632
|
4.772
|
NM_175451
|
Ckap4
|
cytoskeleton-associated protein 4
|
|
chr6_-_37391993
|
4.764
|
NM_178661
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2
|
|
chr18_-_67884185
|
4.714
|
NM_001127177
NM_008977
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2
|
|
chrX_+_70962097
|
4.684
|
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr14_-_47896464
|
4.655
|
|
Wdhd1
|
WD repeat and HMG-box DNA binding protein 1
|
|
chr5_-_111082269
|
4.644
|
|
Noc4l
|
nucleolar complex associated 4 homolog (S. cerevisiae)
|
|
chr1_-_52557130
|
4.627
|
NM_008667
|
Nab1
|
Ngfi-A binding protein 1
|
|
chr10_-_127962914
|
4.625
|
NM_011843
|
Esyt1
|
extended synaptotagmin-like protein 1
|
|
chr12_-_74387687
|
4.622
|
NM_029580
|
Trmt5
|
TRM5 tRNA methyltransferase 5 homolog (S. cerevisiae)
|
|
chr2_+_35991917
|
4.585
|
NM_026422
|
Mrrf
|
mitochondrial ribosome recycling factor
|
|
chrX_+_70962057
|
4.576
|
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1
|
|
chr10_-_127613623
|
4.575
|
NM_019711
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr7_+_109358966
|
4.561
|
|
Pgap2
|
post-GPI attachment to proteins 2
|
|
chr8_-_82235601
|
4.554
|
NM_015751
|
Abce1
|
ATP-binding cassette, sub-family E (OABP), member 1
|
|
chrX_-_91368427
|
4.546
|
|
Zfx
|
zinc finger protein X-linked
|
|
chr19_+_43764161
|
4.535
|
NM_053103
|
Entpd7
|
ectonucleoside triphosphate diphosphohydrolase 7
|
|
chr2_-_25221101
|
4.526
|
NM_001033293
|
Uap1l1
|
UDP-N-acteylglucosamine pyrophosphorylase 1-like 1
|
|
chr6_+_140572056
|
4.508
|
NM_001005605
|
Aebp2
|
AE binding protein 2
|
|
chr11_-_69734838
|
4.499
|
NM_001166589
NM_001166591
NM_181582
|
Eif5a
|
eukaryotic translation initiation factor 5A
|
|
chr4_+_151413427
|
4.497
|
NM_001159599
NM_028727
|
Nol9
|
nucleolar protein 9
|
|
chr4_-_150432002
|
4.471
|
NM_009498
|
Vamp3
|
vesicle-associated membrane protein 3
|
|
chr15_-_27560282
|
4.460
|
NM_001013792
|
Fam105b
|
family with sequence similarity 105, member B
|
|
chr16_+_57549340
|
4.446
|
NM_001177871
|
Filip1l
|
filamin A interacting protein 1-like
|
|
chr2_-_119980243
|
4.433
|
NM_133838
|
Ehd4
|
EH-domain containing 4
|
|
chr10_-_29920326
|
4.429
|
NM_001109747
|
Cenpw
|
centromere protein W
|
|
chr2_-_132078887
|
4.409
|
NM_011045
|
Pcna
|
proliferating cell nuclear antigen
|
|
chr8_+_74659107
|
4.393
|
NM_001001491
|
Tpm4
|
tropomyosin 4
|
|
chr2_+_32631305
|
4.355
|
NM_178595
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae)
|
|
chr10_-_83996516
|
4.341
|
|
Ckap4
|
cytoskeleton-associated protein 4
|
|
chr14_-_87540920
|
4.297
|
NM_019670
|
Diap3
|
diaphanous homolog 3 (Drosophila)
|
|
chr8_-_108535317
|
4.286
|
NM_028038
|
Ddx28
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 28
|
|
chr9_-_85220730
|
4.283
|
NM_001160378
NM_001160379
|
Fam46a
|
family with sequence similarity 46, member A
|
|
chr2_-_119980210
|
4.270
|
|
Ehd4
|
EH-domain containing 4
|
|
chr19_+_7568529
|
4.263
|
NM_001163505
|
Atl3
|
atlastin GTPase 3
|
|
chr8_+_108424267
|
4.196
|
NM_173432
|
Pskh1
|
protein serine kinase H1
|
|
chr17_-_26206487
|
4.181
|
|
Rab11fip3
|
RAB11 family interacting protein 3 (class II)
|
|
chr9_+_113650560
|
4.130
|
NM_001114347
|
Clasp2
|
CLIP associating protein 2
|
|
chr8_+_77633426
|
4.097
|
NM_008566
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae)
|
|
chr10_-_53795556
|
4.096
|
NM_008548
|
Man1a
|
mannosidase 1, alpha
|
|
chr1_+_52065469
|
4.083
|
|
Stat4
|
signal transducer and activator of transcription 4
|
|
chr11_-_86621130
|
4.081
|
NM_026191
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr19_+_37450854
|
4.051
|
NM_010615
|
Kif11
|
kinesin family member 11
|
|
chr15_-_81230036
|
4.050
|
NM_133726
|
St13
|
suppression of tumorigenicity 13
|
|
chr7_+_16894925
|
4.029
|
NM_133234
|
Bbc3
|
BCL2 binding component 3
|
|
chr11_-_69734709
|
4.001
|
NM_001166595
|
Eif5a
|
eukaryotic translation initiation factor 5A
|
|
chr2_+_164571458
|
4.001
|
NM_133763
|
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1
|
|
chr7_+_31338319
|
3.998
|
NM_001012401
|
Hspb6
|
heat shock protein, alpha-crystallin-related, B6
|
|
chr17_+_15841889
|
3.976
|
NM_007690
|
Chd1
|
chromodomain helicase DNA binding protein 1
|
|
chr5_+_76569257
|
3.968
|
NM_020611
|
Srd5a3
|
steroid 5 alpha-reductase 3
|
|
chr8_+_41597112
|
3.955
|
NM_033560
|
Vps37a
|
vacuolar protein sorting 37A (yeast)
|
|
chr13_-_46823191
|
3.946
|
NM_175749
|
Nup153
|
nucleoporin 153
|
|
chr12_+_111976121
|
3.931
|
NM_027149
|
Wdr20a
|
WD repeat domain 20A
|
|
chr19_+_5637712
|
3.930
|
|
Rela
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr13_+_73397944
|
3.929
|
NM_018885
|
Irx4
|
Iroquois related homeobox 4 (Drosophila)
|
|
chr1_+_95375503
|
3.918
|
NM_001159717
NM_001159718
NM_010891
NM_001159719
|
Sept2
|
septin 2
|
|
chr12_+_110031153
|
3.900
|
|
Yy1
|
YY1 transcription factor
|
|
chr8_+_34842304
|
3.898
|
NM_001167921
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit)
|
|
chr10_+_51742333
|
3.891
|
|
Vgll2
|
vestigial like 2 homolog (Drosophila)
|
|
chr5_-_138227888
|
3.871
|
NM_144913
|
Mepce
|
methylphosphate capping enzyme
|
|
chr13_+_73398198
|
3.871
|
|
Irx4
|
Iroquois related homeobox 4 (Drosophila)
|
|
chr6_+_140572347
|
3.855
|
|
Aebp2
|
AE binding protein 2
|
|
chr10_+_126478969
|
3.830
|
NM_010792
|
Mettl1
|
methyltransferase like 1
|
|
chr2_-_76511070
|
3.818
|
NM_010222
|
Fkbp7
|
FK506 binding protein 7
|
|
chr19_+_5637741
|
3.786
|
|
Rela
|
v-rel reticuloendotheliosis viral oncogene homolog A (avian)
|
|
chr2_+_125684938
|
3.779
|
NM_175154
|
Galk2
|
galactokinase 2
|
|
chr8_+_108535363
|
3.775
|
NM_025518
|
Dus2l
|
dihydrouridine synthase 2-like (SMM1, S. cerevisiae)
|
|
chr2_+_25110946
|
3.766
|
NM_177344
|
Tmem203
|
transmembrane protein 203
|
|
chr11_-_115992196
|
3.761
|
NM_178802
|
Trim65
|
tripartite motif-containing 65
|
|
chr11_+_120810346
|
3.736
|
NM_001038653
NM_001038654
|
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3
|
|
chr15_+_34012606
|
3.726
|
|
Mtdh
|
metadherin
|
|
chr2_+_31829969
|
3.719
|
NM_172268
|
Nup214
|
nucleoporin 214
|
|
chrX_-_138825032
|
3.715
|
NM_001033600
NM_019477
NM_207625
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr3_-_8923789
|
3.705
|
NM_025434
|
Mrps28
|
mitochondrial ribosomal protein S28
|
|
chr9_+_48303449
|
3.702
|
NM_001004191
|
Gm5617
|
predicted gene 5617
|
|
chr8_-_72498125
|
3.702
|
|
Gatad2a
|
GATA zinc finger domain containing 2A
|
|
chr1_+_82313139
|
3.699
|
NM_029777
|
Rhbdd1
|
rhomboid domain containing 1
|
|
chr7_+_54101157
|
3.693
|
NM_010699
|
Ldha
|
lactate dehydrogenase A
|
|
chr19_-_34549624
|
3.693
|
NM_009890
|
Ch25h
|
cholesterol 25-hydroxylase
|
|
chr8_-_107155195
|
3.681
|
NM_019662
|
Rrad
|
Ras-related associated with diabetes
|
|
chr2_-_103601232
|
3.676
|
NM_153126
|
Nat10
|
N-acetyltransferase 10
|
|
chr9_+_108462759
|
3.662
|
NM_011830
|
Impdh2
|
inosine 5'-phosphate dehydrogenase 2
|
|
chr10_+_110910277
|
3.654
|
|
Nap1l1
|
nucleosome assembly protein 1-like 1
|
|
chr17_+_35118956
|
3.632
|
NM_030597
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae)
|
|
chr9_+_107951572
|
3.591
|
NM_177910
|
Gmppb
|
GDP-mannose pyrophosphorylase B
|
|
chr11_+_53164267
|
3.590
|
NM_033565
|
Aff4
|
AF4/FMR2 family, member 4
|
|
chr14_+_64479129
|
3.566
|
NM_028228
|
Pinx1
|
PIN2/TERF1 interacting, telomerase inhibitor 1
|
|
chr17_+_34987323
|
3.563
|
NM_001045863
NM_001045864
NM_138580
|
Rdbp
|
RD RNA-binding protein
|
|
chr14_-_32064403
|
3.546
|
NM_027949
|
Phf7
|
PHD finger protein 7
|
|
chr5_+_48763568
|
3.544
|
NM_025755
|
Pacrgl
|
PARK2 co-regulated-like
|
|
chr18_+_66618186
|
3.526
|
NM_021451
|
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1
|
|
chr7_-_31514527
|
3.520
|
NM_027215
|
Tmem147
|
transmembrane protein 147
|
|
chr15_-_59205688
|
3.518
|
NM_153548
|
E430025E21Rik
|
RIKEN cDNA E430025E21 gene
|
|
chrX_-_35917684
|
3.516
|
|
Cul4b
|
cullin 4B
|
|
chr1_-_97564064
|
3.510
|
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|