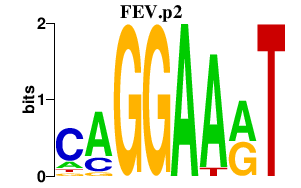
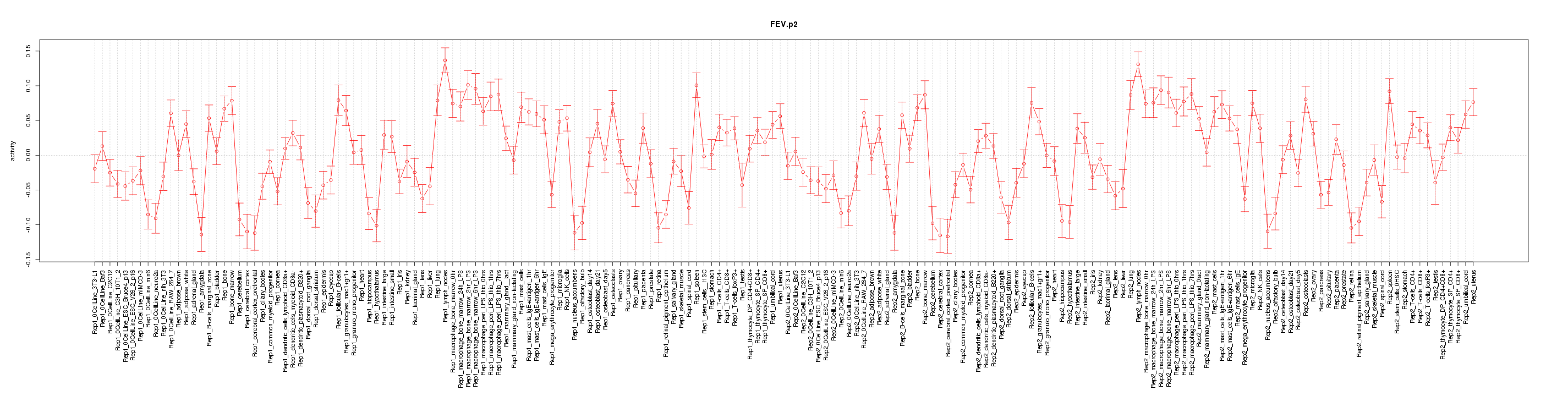
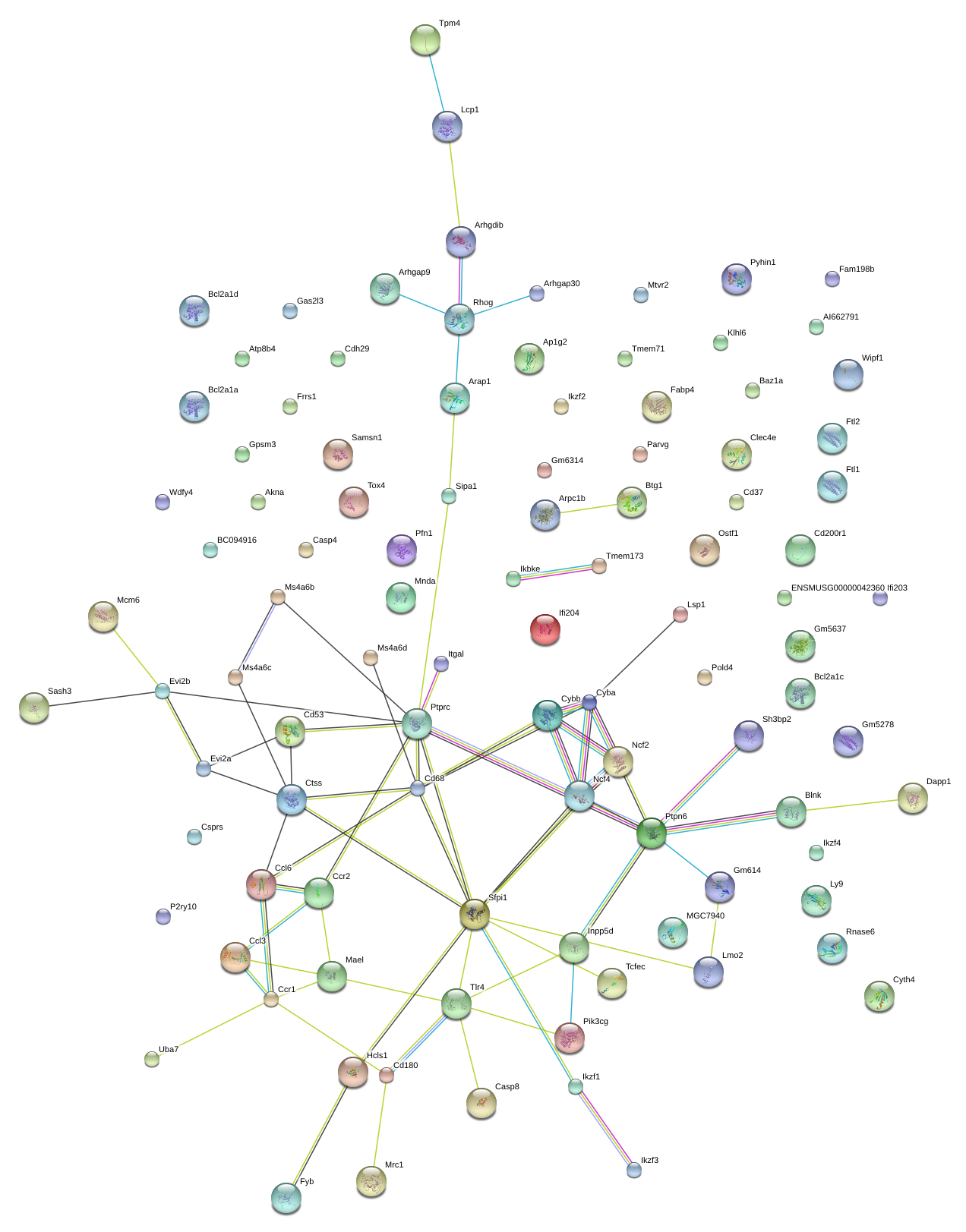
|
1.41
|
1.86e-40
|
GO:0044260
|
cellular macromolecule metabolic process
|
|
1.37
|
7.95e-39
|
GO:0043170
|
macromolecule metabolic process
|
|
1.28
|
4.27e-30
|
GO:0044238
|
primary metabolic process
|
|
1.28
|
2.30e-29
|
GO:0044237
|
cellular metabolic process
|
|
1.24
|
9.99e-26
|
GO:0008152
|
metabolic process
|
|
1.85
|
5.54e-25
|
GO:0002376
|
immune system process
|
|
1.47
|
2.77e-24
|
GO:0010467
|
gene expression
|
|
1.33
|
7.37e-22
|
GO:0019222
|
regulation of metabolic process
|
|
1.38
|
2.73e-21
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.42
|
5.84e-21
|
GO:0090304
|
nucleic acid metabolic process
|
|
1.35
|
1.68e-20
|
GO:0031323
|
regulation of cellular metabolic process
|
|
1.44
|
2.84e-19
|
GO:0016070
|
RNA metabolic process
|
|
1.34
|
2.93e-19
|
GO:0080090
|
regulation of primary metabolic process
|
|
1.34
|
3.08e-18
|
GO:0060255
|
regulation of macromolecule metabolic process
|
|
1.52
|
2.57e-17
|
GO:0048583
|
regulation of response to stimulus
|
|
1.43
|
3.74e-17
|
GO:0044267
|
cellular protein metabolic process
|
|
1.41
|
5.06e-17
|
GO:0009059
|
macromolecule biosynthetic process
|
|
1.31
|
1.29e-16
|
GO:0006807
|
nitrogen compound metabolic process
|
|
1.40
|
1.40e-16
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
|
1.37
|
2.18e-16
|
GO:0048518
|
positive regulation of biological process
|
|
1.32
|
3.25e-16
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
|
1.35
|
1.40e-15
|
GO:0071840
|
cellular component organization or biogenesis
|
|
1.36
|
2.74e-15
|
GO:0051171
|
regulation of nitrogen compound metabolic process
|
|
1.36
|
3.11e-15
|
GO:0019538
|
protein metabolic process
|
|
1.36
|
5.35e-15
|
GO:0019219
|
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.37
|
1.08e-14
|
GO:0010556
|
regulation of macromolecule biosynthetic process
|
|
1.37
|
1.35e-14
|
GO:0048522
|
positive regulation of cellular process
|
|
2.17
|
2.47e-14
|
GO:0001775
|
cell activation
|
|
1.35
|
2.97e-14
|
GO:0010468
|
regulation of gene expression
|
|
2.26
|
3.15e-14
|
GO:0045321
|
leukocyte activation
|
|
1.35
|
3.57e-14
|
GO:0009889
|
regulation of biosynthetic process
|
|
1.34
|
9.72e-14
|
GO:0031326
|
regulation of cellular biosynthetic process
|
|
1.35
|
3.51e-13
|
GO:2000112
|
regulation of cellular macromolecule biosynthetic process
|
|
1.11
|
3.52e-13
|
GO:0009987
|
cellular process
|
|
1.30
|
6.95e-13
|
GO:0009058
|
biosynthetic process
|
|
1.31
|
8.65e-13
|
GO:0044249
|
cellular biosynthetic process
|
|
1.77
|
1.02e-12
|
GO:0002682
|
regulation of immune system process
|
|
1.94
|
5.55e-12
|
GO:0002684
|
positive regulation of immune system process
|
|
1.95
|
8.23e-12
|
GO:0006955
|
immune response
|
|
1.35
|
1.21e-11
|
GO:0051252
|
regulation of RNA metabolic process
|
|
1.31
|
1.99e-11
|
GO:0016043
|
cellular component organization
|
|
1.46
|
2.30e-11
|
GO:0009893
|
positive regulation of metabolic process
|
|
1.43
|
2.65e-11
|
GO:0006950
|
response to stress
|
|
1.35
|
2.87e-11
|
GO:0006355
|
regulation of transcription, DNA-dependent
|
|
1.48
|
5.23e-11
|
GO:0010604
|
positive regulation of macromolecule metabolic process
|
|
1.33
|
1.43e-10
|
GO:0048519
|
negative regulation of biological process
|
|
1.45
|
4.35e-10
|
GO:0031325
|
positive regulation of cellular metabolic process
|
|
2.18
|
8.26e-10
|
GO:0046649
|
lymphocyte activation
|
|
1.92
|
1.69e-09
|
GO:0050776
|
regulation of immune response
|
|
1.34
|
1.74e-09
|
GO:0048523
|
negative regulation of cellular process
|
|
1.59
|
1.77e-09
|
GO:0048584
|
positive regulation of response to stimulus
|
|
1.50
|
1.83e-09
|
GO:0035556
|
intracellular signal transduction
|
|
1.38
|
1.91e-09
|
GO:0032774
|
RNA biosynthetic process
|
|
1.33
|
2.16e-09
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
|
1.38
|
2.27e-09
|
GO:0006351
|
transcription, DNA-dependent
|
|
1.39
|
2.45e-09
|
GO:0006464
|
protein modification process
|
|
1.71
|
4.58e-09
|
GO:0006396
|
RNA processing
|
|
1.46
|
5.17e-09
|
GO:0009966
|
regulation of signal transduction
|
|
1.37
|
6.85e-09
|
GO:0043412
|
macromolecule modification
|
|
2.40
|
8.33e-09
|
GO:0002764
|
immune response-regulating signaling pathway
|
|
1.41
|
8.62e-09
|
GO:0006996
|
organelle organization
|
|
1.49
|
1.15e-08
|
GO:0010941
|
regulation of cell death
|
|
1.50
|
1.23e-08
|
GO:0042981
|
regulation of apoptosis
|
|
1.49
|
2.69e-08
|
GO:0043067
|
regulation of programmed cell death
|
|
2.27
|
3.42e-08
|
GO:0002521
|
leukocyte differentiation
|
|
1.51
|
3.67e-08
|
GO:0042127
|
regulation of cell proliferation
|
|
1.99
|
8.72e-08
|
GO:0050778
|
positive regulation of immune response
|
|
1.47
|
2.43e-07
|
GO:0006357
|
regulation of transcription from RNA polymerase II promoter
|
|
2.31
|
2.80e-07
|
GO:0002757
|
immune response-activating signal transduction
|
|
1.66
|
3.00e-07
|
GO:0010942
|
positive regulation of cell death
|
|
1.52
|
3.24e-07
|
GO:0007049
|
cell cycle
|
|
2.12
|
3.42e-07
|
GO:0022613
|
ribonucleoprotein complex biogenesis
|
|
1.67
|
4.06e-07
|
GO:0043068
|
positive regulation of programmed cell death
|
|
1.67
|
4.25e-07
|
GO:0043065
|
positive regulation of apoptosis
|
|
1.55
|
4.44e-07
|
GO:0016265
|
death
|
|
1.55
|
5.76e-07
|
GO:0008219
|
cell death
|
|
1.78
|
7.97e-07
|
GO:0016071
|
mRNA metabolic process
|
|
3.02
|
8.32e-07
|
GO:0002768
|
immune response-regulating cell surface receptor signaling pathway
|
|
1.84
|
1.81e-06
|
GO:0030097
|
hemopoiesis
|
|
1.47
|
1.87e-06
|
GO:0010557
|
positive regulation of macromolecule biosynthetic process
|
|
2.04
|
1.90e-06
|
GO:0071843
|
cellular component biogenesis at cellular level
|
|
1.79
|
1.93e-06
|
GO:0048534
|
hemopoietic or lymphoid organ development
|
|
2.24
|
3.52e-06
|
GO:0042110
|
T cell activation
|
|
1.44
|
3.96e-06
|
GO:0009891
|
positive regulation of biosynthetic process
|
|
1.44
|
4.24e-06
|
GO:0031328
|
positive regulation of cellular biosynthetic process
|
|
1.87
|
4.40e-06
|
GO:0001817
|
regulation of cytokine production
|
|
1.55
|
4.54e-06
|
GO:0006915
|
apoptosis
|
|
2.09
|
4.75e-06
|
GO:0002253
|
activation of immune response
|
|
1.42
|
5.40e-06
|
GO:0008104
|
protein localization
|
|
2.27
|
6.13e-06
|
GO:0042254
|
ribosome biogenesis
|
|
1.48
|
9.32e-06
|
GO:0015031
|
protein transport
|
|
1.73
|
9.78e-06
|
GO:0002520
|
immune system development
|
|
1.27
|
1.06e-05
|
GO:0071842
|
cellular component organization at cellular level
|
|
1.53
|
1.08e-05
|
GO:0012501
|
programmed cell death
|
|
1.56
|
1.10e-05
|
GO:0022402
|
cell cycle process
|
|
1.63
|
1.20e-05
|
GO:0080134
|
regulation of response to stress
|
|
1.40
|
1.33e-05
|
GO:0009892
|
negative regulation of metabolic process
|
|
1.47
|
1.35e-05
|
GO:0045184
|
establishment of protein localization
|
|
1.68
|
1.43e-05
|
GO:0009611
|
response to wounding
|
|
1.41
|
1.75e-05
|
GO:0044085
|
cellular component biogenesis
|
|
1.41
|
1.82e-05
|
GO:0010605
|
negative regulation of macromolecule metabolic process
|
|
1.45
|
1.96e-05
|
GO:0010628
|
positive regulation of gene expression
|
|
2.89
|
2.28e-05
|
GO:0002429
|
immune response-activating cell surface receptor signaling pathway
|
|
1.58
|
2.71e-05
|
GO:0006952
|
defense response
|
|
3.09
|
3.72e-05
|
GO:0030888
|
regulation of B cell proliferation
|
|
1.51
|
4.06e-05
|
GO:0070887
|
cellular response to chemical stimulus
|
|
1.80
|
4.29e-05
|
GO:0031347
|
regulation of defense response
|
|
1.67
|
4.41e-05
|
GO:0010627
|
regulation of intracellular protein kinase cascade
|
|
1.77
|
4.89e-05
|
GO:0006397
|
mRNA processing
|
|
1.56
|
5.11e-05
|
GO:0008284
|
positive regulation of cell proliferation
|
|
1.85
|
5.88e-05
|
GO:0002694
|
regulation of leukocyte activation
|
|
1.36
|
6.37e-05
|
GO:0033036
|
macromolecule localization
|
|
2.57
|
6.38e-05
|
GO:0050864
|
regulation of B cell activation
|
|
2.19
|
7.24e-05
|
GO:0030098
|
lymphocyte differentiation
|
|
1.78
|
7.87e-05
|
GO:0008380
|
RNA splicing
|
|
1.32
|
8.87e-05
|
GO:0023051
|
regulation of signaling
|
|
1.59
|
9.33e-05
|
GO:0051726
|
regulation of cell cycle
|
|
2.96
|
1.19e-04
|
GO:0050851
|
antigen receptor-mediated signaling pathway
|
|
2.18
|
1.19e-04
|
GO:0050670
|
regulation of lymphocyte proliferation
|
|
2.10
|
1.28e-04
|
GO:0001819
|
positive regulation of cytokine production
|
|
1.87
|
1.41e-04
|
GO:0006954
|
inflammatory response
|
|
2.16
|
1.61e-04
|
GO:0032944
|
regulation of mononuclear cell proliferation
|
|
1.64
|
1.73e-04
|
GO:0000278
|
mitotic cell cycle
|
|
2.14
|
1.77e-04
|
GO:0048872
|
homeostasis of number of cells
|
|
1.38
|
1.98e-04
|
GO:0031324
|
negative regulation of cellular metabolic process
|
|
1.80
|
1.98e-04
|
GO:0050865
|
regulation of cell activation
|
|
2.33
|
2.02e-04
|
GO:0016072
|
rRNA metabolic process
|
|
2.97
|
2.06e-04
|
GO:0007249
|
I-kappaB kinase/NF-kappaB cascade
|
|
1.84
|
2.14e-04
|
GO:0030036
|
actin cytoskeleton organization
|
|
2.34
|
2.40e-04
|
GO:0006364
|
rRNA processing
|
|
1.44
|
2.44e-04
|
GO:0045893
|
positive regulation of transcription, DNA-dependent
|
|
1.40
|
2.53e-04
|
GO:0051173
|
positive regulation of nitrogen compound metabolic process
|
|
1.86
|
2.69e-04
|
GO:0051249
|
regulation of lymphocyte activation
|
|
2.13
|
2.89e-04
|
GO:0070663
|
regulation of leukocyte proliferation
|
|
1.96
|
3.04e-04
|
GO:0002697
|
regulation of immune effector process
|
|
1.80
|
3.11e-04
|
GO:0030029
|
actin filament-based process
|
|
1.37
|
3.12e-04
|
GO:0006793
|
phosphorus metabolic process
|
|
2.79
|
3.33e-04
|
GO:0050871
|
positive regulation of B cell activation
|
|
1.66
|
3.46e-04
|
GO:0031401
|
positive regulation of protein modification process
|
|
1.27
|
3.50e-04
|
GO:0065008
|
regulation of biological quality
|
|
1.74
|
3.62e-04
|
GO:0007264
|
small GTPase mediated signal transduction
|
|
1.27
|
3.73e-04
|
GO:0048513
|
organ development
|
|
1.37
|
4.02e-04
|
GO:0006796
|
phosphate metabolic process
|
|
1.99
|
4.37e-04
|
GO:0002252
|
immune effector process
|
|
1.42
|
4.73e-04
|
GO:0051254
|
positive regulation of RNA metabolic process
|
|
1.59
|
4.90e-04
|
GO:0032583
|
regulation of gene-specific transcription
|
|
2.32
|
6.82e-04
|
GO:0006414
|
translational elongation
|
|
1.81
|
7.49e-04
|
GO:0010564
|
regulation of cell cycle process
|
|
1.76
|
7.97e-04
|
GO:0051056
|
regulation of small GTPase mediated signal transduction
|
|
1.39
|
8.13e-04
|
GO:0045935
|
positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
1.81
|
9.30e-04
|
GO:0046578
|
regulation of Ras protein signal transduction
|
|
1.94
|
9.32e-04
|
GO:0031349
|
positive regulation of defense response
|
|
1.57
|
9.99e-04
|
GO:0032270
|
positive regulation of cellular protein metabolic process
|
|
2.21
|
1.05e-03
|
GO:0032103
|
positive regulation of response to external stimulus
|
|
1.20
|
1.21e-03
|
GO:0048856
|
anatomical structure development
|
|
1.44
|
1.43e-03
|
GO:0033554
|
cellular response to stress
|
|
1.29
|
1.44e-03
|
GO:0065009
|
regulation of molecular function
|
|
1.95
|
1.78e-03
|
GO:0002696
|
positive regulation of leukocyte activation
|
|
1.57
|
1.81e-03
|
GO:0006974
|
response to DNA damage stimulus
|
|
1.55
|
1.89e-03
|
GO:0051247
|
positive regulation of protein metabolic process
|
|
2.30
|
1.93e-03
|
GO:0030217
|
T cell differentiation
|
|
1.47
|
2.03e-03
|
GO:0045944
|
positive regulation of transcription from RNA polymerase II promoter
|
|
1.09
|
2.15e-03
|
GO:0065007
|
biological regulation
|
|
1.35
|
2.66e-03
|
GO:0032268
|
regulation of cellular protein metabolic process
|
|
1.91
|
2.88e-03
|
GO:0050867
|
positive regulation of cell activation
|
|
1.21
|
3.47e-03
|
GO:0048731
|
system development
|
|
1.34
|
3.74e-03
|
GO:0032879
|
regulation of localization
|
|
1.70
|
4.16e-03
|
GO:0010608
|
posttranscriptional regulation of gene expression
|
|
1.49
|
4.19e-03
|
GO:0051276
|
chromosome organization
|
|
1.95
|
4.22e-03
|
GO:0051251
|
positive regulation of lymphocyte activation
|
|
1.09
|
4.51e-03
|
GO:0050789
|
regulation of biological process
|
|
2.17
|
4.73e-03
|
GO:0002699
|
positive regulation of immune effector process
|
|
1.77
|
4.81e-03
|
GO:0034097
|
response to cytokine stimulus
|
|
1.17
|
4.90e-03
|
GO:0051179
|
localization
|
|
1.40
|
4.99e-03
|
GO:0010558
|
negative regulation of macromolecule biosynthetic process
|
|
1.17
|
5.35e-03
|
GO:0032502
|
developmental process
|
|
1.38
|
5.43e-03
|
GO:0044093
|
positive regulation of molecular function
|
|
2.03
|
5.59e-03
|
GO:0050727
|
regulation of inflammatory response
|
|
1.38
|
6.11e-03
|
GO:0009890
|
negative regulation of biosynthetic process
|
|
1.58
|
6.24e-03
|
GO:0007243
|
intracellular protein kinase cascade
|
|
1.58
|
6.24e-03
|
GO:0023014
|
signal transduction via phosphorylation event
|
|
1.58
|
8.07e-03
|
GO:0051301
|
cell division
|
|
1.71
|
8.81e-03
|
GO:0043193
|
positive regulation of gene-specific transcription
|
|
1.51
|
8.86e-03
|
GO:0016044
|
cellular membrane organization
|
|
2.26
|
9.08e-03
|
GO:0042035
|
regulation of cytokine biosynthetic process
|
|
1.24
|
9.50e-03
|
GO:0042221
|
response to chemical stimulus
|
|
2.54
|
9.67e-03
|
GO:0002274
|
myeloid leukocyte activation
|
|
2.95
|
9.87e-03
|
GO:0001776
|
leukocyte homeostasis
|
|
1.51
|
1.06e-02
|
GO:0022403
|
cell cycle phase
|
|
1.51
|
1.14e-02
|
GO:0061024
|
membrane organization
|
|
2.11
|
1.14e-02
|
GO:0002703
|
regulation of leukocyte mediated immunity
|
|
1.69
|
1.15e-02
|
GO:0032880
|
regulation of protein localization
|
|
2.67
|
1.37e-02
|
GO:0002687
|
positive regulation of leukocyte migration
|
|
1.09
|
1.38e-02
|
GO:0050794
|
regulation of cellular process
|
|
1.63
|
1.58e-02
|
GO:0031329
|
regulation of cellular catabolic process
|
|
1.35
|
1.61e-02
|
GO:0016310
|
phosphorylation
|
|
2.53
|
1.67e-02
|
GO:0042108
|
positive regulation of cytokine biosynthetic process
|
|
2.95
|
1.72e-02
|
GO:0002260
|
lymphocyte homeostasis
|
|
2.95
|
1.72e-02
|
GO:0030890
|
positive regulation of B cell proliferation
|
|
1.39
|
1.74e-02
|
GO:0010629
|
negative regulation of gene expression
|
|
1.48
|
1.77e-02
|
GO:0043066
|
negative regulation of apoptosis
|
|
1.37
|
1.77e-02
|
GO:0031327
|
negative regulation of cellular biosynthetic process
|
|
2.17
|
1.92e-02
|
GO:0050671
|
positive regulation of lymphocyte proliferation
|
|
1.47
|
2.08e-02
|
GO:0043069
|
negative regulation of programmed cell death
|
|
1.60
|
2.12e-02
|
GO:0006917
|
induction of apoptosis
|
|
2.45
|
2.14e-02
|
GO:0002263
|
cell activation involved in immune response
|
|
2.45
|
2.14e-02
|
GO:0002366
|
leukocyte activation involved in immune response
|
|
1.55
|
2.26e-02
|
GO:0010551
|
regulation of gene-specific transcription from RNA polymerase II promoter
|
|
2.18
|
2.34e-02
|
GO:0042113
|
B cell activation
|
|
1.46
|
2.39e-02
|
GO:0009967
|
positive regulation of signal transduction
|
|
1.60
|
2.45e-02
|
GO:0012502
|
induction of programmed cell death
|
|
2.48
|
2.49e-02
|
GO:0032675
|
regulation of interleukin-6 production
|
|
1.74
|
2.54e-02
|
GO:0030155
|
regulation of cell adhesion
|
|
2.14
|
2.58e-02
|
GO:0032946
|
positive regulation of mononuclear cell proliferation
|
|
2.72
|
2.61e-02
|
GO:0043409
|
negative regulation of MAPKKK cascade
|
|
2.56
|
3.29e-02
|
GO:0050729
|
positive regulation of inflammatory response
|
|
1.45
|
3.71e-02
|
GO:0060548
|
negative regulation of cell death
|
|
1.55
|
3.77e-02
|
GO:0001944
|
vasculature development
|
|
2.34
|
3.79e-02
|
GO:0060326
|
cell chemotaxis
|
|
1.30
|
3.90e-02
|
GO:0051641
|
cellular localization
|
|
1.28
|
3.91e-02
|
GO:0050790
|
regulation of catalytic activity
|
|
1.37
|
4.00e-02
|
GO:0006468
|
protein phosphorylation
|
|
1.29
|
4.07e-02
|
GO:0051246
|
regulation of protein metabolic process
|
|
1.68
|
4.42e-02
|
GO:0043408
|
regulation of MAPKKK cascade
|
|
1.98
|
4.52e-02
|
GO:0043122
|
regulation of I-kappaB kinase/NF-kappaB cascade
|
|
2.09
|
4.57e-02
|
GO:0070665
|
positive regulation of leukocyte proliferation
|
|
1.20
|
4.84e-02
|
GO:0048869
|
cellular developmental process
|
|
1.58
|
4.89e-02
|
GO:0018193
|
peptidyl-amino acid modification
|
|
1.54
|
4.92e-02
|
GO:0016568
|
chromatin modification
|