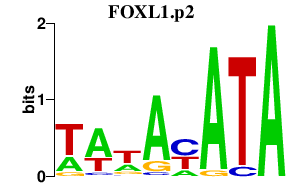
|
chr13_+_49704471
|
35.230
|
|
Ogn
|
osteoglycin
|
|
chr1_+_45402907
|
34.804
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr10_+_97028134
|
29.635
|
NM_008524
|
Lum
|
lumican
|
|
chr10_+_97028409
|
27.064
|
|
Lum
|
lumican
|
|
chr10_+_97028456
|
25.576
|
|
Lum
|
lumican
|
|
chr17_-_46155601
|
25.212
|
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr7_-_125629996
|
24.952
|
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr19_+_9092973
|
24.576
|
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin)
|
|
chr6_-_14705104
|
23.724
|
NM_080464
|
Ppp1r3a
|
protein phosphatase 1, regulatory (inhibitor) subunit 3A
|
|
chr6_+_4465627
|
22.035
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr1_+_92100572
|
21.972
|
NM_007722
|
Cxcr7
|
chemokine (C-X-C motif) receptor 7
|
|
chr16_+_45093784
|
21.497
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr14_+_102239673
|
21.398
|
|
Lmo7
|
LIM domain only 7
|
|
chr1_+_59543450
|
21.129
|
|
|
|
|
chr16_-_85862579
|
19.548
|
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr13_+_49639847
|
19.540
|
|
Aspn
|
asporin
|
|
chr13_+_49639799
|
18.274
|
NM_001172481
NM_025711
|
Aspn
|
asporin
|
|
chr14_-_104213981
|
17.953
|
|
|
|
|
chr8_+_55635683
|
15.470
|
NM_029569
|
Asb5
|
ankyrin repeat and SOCs box-containing 5
|
|
chr11_+_67013553
|
15.243
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chrX_+_130385690
|
15.073
|
|
Tnmd
|
tenomodulin
|
|
chr3_+_108163704
|
14.137
|
|
Sort1
|
sortilin 1
|
|
chr5_-_69933835
|
13.998
|
NM_023784
|
Yipf7
|
Yip1 domain family, member 7
|
|
chr2_-_126719808
|
13.692
|
|
2010106G01Rik
|
RIKEN cDNA 2010106G01 gene
|
|
chr1_-_45559904
|
13.535
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr3_+_95303177
|
13.477
|
NM_007802
|
Ctsk
|
cathepsin K
|
|
chr6_+_15750966
|
13.179
|
|
Mdfic
|
MyoD family inhibitor domain containing
|
|
chr2_-_173053287
|
12.887
|
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1
|
|
chrX_+_130385546
|
12.843
|
NM_022322
|
Tnmd
|
tenomodulin
|
|
chr8_+_55635768
|
12.828
|
|
Asb5
|
ankyrin repeat and SOCs box-containing 5
|
|
chr16_+_56477956
|
12.699
|
NM_001014399
NM_001014422
NM_001014423
NM_001014424
NM_178790
|
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein
|
|
chr1_+_97230610
|
12.577
|
|
Fam174a
|
family with sequence similarity 174, member A
|
|
chr4_+_57907032
|
12.403
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr4_+_57907600
|
12.403
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr16_-_76288569
|
12.360
|
|
Nrip1
|
nuclear receptor interacting protein 1
|
|
chr4_+_57906588
|
12.291
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr16_+_45093816
|
12.210
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr5_-_124028994
|
11.976
|
|
Clip1
|
CAP-GLY domain containing linker protein 1
|
|
chr1_+_45401659
|
11.861
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr2_-_62411970
|
11.648
|
NM_007986
|
Fap
|
fibroblast activation protein
|
|
chr6_+_55286292
|
11.509
|
NM_007472
|
Aqp1
|
aquaporin 1
|
|
chr9_+_92502076
|
11.491
|
|
|
|
|
chr6_+_4471231
|
11.353
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr19_+_20676453
|
11.202
|
NM_013467
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1
|
|
chr5_-_17394564
|
10.942
|
|
Cd36
|
CD36 antigen
|
|
chr1_-_66991580
|
10.722
|
|
Myl1
|
myosin, light polypeptide 1
|
|
chr13_+_83803830
|
10.686
|
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chrX_+_80194208
|
10.605
|
NM_007868
|
Dmd
|
dystrophin, muscular dystrophy
|
|
chr1_+_171585621
|
10.591
|
NM_009063
|
Rgs5
|
regulator of G-protein signaling 5
|
|
chr9_-_70452528
|
10.550
|
|
Fam63b
|
family with sequence similarity 63, member B
|
|
chr4_-_9570742
|
10.550
|
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr5_-_17341512
|
10.488
|
|
Cd36
|
CD36 antigen
|
|
chr9_-_101101334
|
10.384
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr15_-_3421486
|
10.210
|
NM_001048147
|
Ghr
|
growth hormone receptor
|
|
chr1_-_45559700
|
10.086
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr3_+_123008796
|
9.955
|
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae)
|
|
chr4_+_48676385
|
9.747
|
NM_026509
|
Murc
|
muscle-related coiled-coil protein
|
|
chr16_-_4523061
|
9.531
|
|
Srl
|
sarcalumenin
|
|
chr2_+_145647868
|
9.500
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr1_-_72905107
|
9.481
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr8_+_15057652
|
9.257
|
NM_008664
|
Myom2
|
myomesin 2
|
|
chr16_+_34995000
|
9.169
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr8_-_25549415
|
9.037
|
NM_026931
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene
|
|
chr3_-_10208575
|
8.908
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr5_+_90889937
|
8.883
|
|
Alb
|
albumin
|
|
chr7_-_31839825
|
8.860
|
NM_052991
NM_194321
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr3_-_27316628
|
8.759
|
|
Fndc3b
|
fibronectin type III domain containing 3B
|
|
chr13_+_23830671
|
8.722
|
NM_015786
|
Hist1h1c
|
histone cluster 1, H1c
|
|
chr6_-_136824217
|
8.695
|
|
Mgp
|
matrix Gla protein
|
|
chr7_-_56103389
|
8.537
|
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr6_+_4455686
|
8.496
|
NM_007743
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr8_-_126418628
|
8.456
|
NM_009606
|
Acta1
|
actin, alpha 1, skeletal muscle
|
|
chr1_-_45559866
|
8.373
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr6_+_4484651
|
8.373
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr8_-_37631772
|
8.354
|
|
Dlc1
|
deleted in liver cancer 1
|
|
chr2_+_145068346
|
8.350
|
NM_053195
|
Slc24a3
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3
|
|
chr11_-_40562196
|
8.266
|
|
|
|
|
chr1_-_45560001
|
8.252
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr11_+_98245124
|
8.252
|
NM_011540
|
Tcap
|
titin-cap
|
|
chr19_+_30020236
|
8.242
|
NM_001164724
|
Il33
|
interleukin 33
|
|
chr14_+_120830603
|
8.194
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr1_-_174149989
|
8.084
|
NM_009813
|
Casq1
|
calsequestrin 1
|
|
chr8_-_37631532
|
7.997
|
|
Dlc1
|
deleted in liver cancer 1
|
|
chrX_+_160876032
|
7.929
|
NM_026853
|
Asb11
|
ankyrin repeat and SOCS box-containing 11
|
|
chr19_-_20802015
|
7.759
|
NM_011921
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7
|
|
chr3_-_122737855
|
7.758
|
NM_021503
|
Myoz2
|
myozenin 2
|
|
chr11_+_29642667
|
7.721
|
|
|
|
|
chr1_-_174149963
|
7.697
|
|
Casq1
|
calsequestrin 1
|
|
chr6_-_142650737
|
7.638
|
NM_001044720
NM_011511
NM_021041
NM_021042
|
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr3_+_19869841
|
7.593
|
|
Cp
|
ceruloplasmin
|
|
chr7_+_149627709
|
7.577
|
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr18_+_69843730
|
7.442
|
|
Tcf4
|
transcription factor 4
|
|
chr4_-_9570815
|
7.413
|
NM_001177852
NM_001177853
NM_001177854
NM_133723
|
Asph
|
aspartate-beta-hydroxylase
|
|
chr8_+_36740640
|
7.404
|
|
Mfhas1
|
malignant fibrous histiocytoma amplified sequence 1
|
|
chr16_-_4523051
|
7.350
|
NM_175347
|
Srl
|
sarcalumenin
|
|
chr3_+_51299153
|
7.268
|
|
|
|
|
chr2_+_131763454
|
7.176
|
|
Prnp
|
prion protein
|
|
chr6_-_136824323
|
7.131
|
NM_008597
|
Mgp
|
matrix Gla protein
|
|
chr9_-_103119803
|
7.111
|
|
Trf
|
transferrin
|
|
chr6_-_136824277
|
7.099
|
|
Mgp
|
matrix Gla protein
|
|
chr11_-_20729367
|
7.041
|
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr8_+_131256428
|
7.037
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr16_+_87731736
|
6.909
|
|
Bach1
|
BTB and CNC homology 1
|
|
chr18_-_39572137
|
6.847
|
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1
|
|
chr7_+_149684731
|
6.704
|
NM_001163664
NM_001163665
NM_001163666
NM_001163667
NM_001163668
NM_001163669
NM_001163670
NM_011620
|
Tnnt3
|
troponin T3, skeletal, fast
|
|
chr10_-_106931783
|
6.679
|
NM_008657
|
Myf6
|
myogenic factor 6
|
|
chr9_+_92502908
|
6.620
|
|
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr5_-_17355533
|
6.542
|
NM_001159556
NM_001159557
|
Cd36
|
CD36 antigen
|
|
chr14_+_13073221
|
6.519
|
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G
|
|
chr1_-_72920776
|
6.516
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chrX_+_10173557
|
6.515
|
|
|
|
|
chr1_+_166726827
|
6.508
|
NM_019759
|
Dpt
|
dermatopontin
|
|
chr6_+_34548512
|
6.478
|
|
Cald1
|
caldesmon 1
|
|
chr7_-_56103405
|
6.431
|
NM_001198841
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr5_-_51848420
|
6.241
|
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr3_-_122939013
|
6.233
|
NM_080451
|
Synpo2
|
synaptopodin 2
|
|
chr3_-_83967952
|
6.219
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr13_-_102462584
|
6.208
|
NM_001024955
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|
|
chrX_-_147248815
|
6.196
|
NM_030700
|
Maged2
|
melanoma antigen, family D, 2
|
|
chr19_-_44471675
|
6.136
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr17_+_24869629
|
6.092
|
NM_025425
|
Rpl3l
|
ribosomal protein L3-like
|
|
chr11_-_40562329
|
6.085
|
|
|
|
|
chr11_-_83391984
|
5.943
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr18_-_58169723
|
5.914
|
|
Fbn2
|
fibrillin 2
|
|
chr11_-_83406475
|
5.876
|
NM_009139
|
Ccl6
|
chemokine (C-C motif) ligand 6
|
|
chr11_+_66984528
|
5.821
|
NM_001039545
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult
|
|
chr17_+_44413168
|
5.789
|
|
Clic5
|
chloride intracellular channel 5
|
|
chr10_+_90564953
|
5.781
|
|
Ikbip
|
IKBKB interacting protein
|
|
chr6_-_129422702
|
5.710
|
NM_020008
|
Clec7a
|
C-type lectin domain family 7, member a
|
|
chrX_+_154137193
|
5.654
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr5_+_129602591
|
5.577
|
NM_001081342
|
Gpr133
|
G protein-coupled receptor 133
|
|
chr7_+_149628285
|
5.574
|
NM_009405
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr5_+_90889913
|
5.557
|
NM_009654
|
Alb
|
albumin
|
|
chr9_+_43877511
|
5.552
|
NM_016808
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr3_-_20022409
|
5.525
|
|
Gyg
|
glycogenin
|
|
chr2_+_32480924
|
5.519
|
NM_001198791
|
Ak1
|
adenylate kinase 1
|
|
chr10_+_96944961
|
5.516
|
NM_007833
|
Dcn
|
decorin
|
|
chr14_+_27707789
|
5.516
|
NM_001170748
NM_080856
|
Asb14
|
ankyrin repeat and SOCS box-containing 14
|
|
chr2_+_69508174
|
5.475
|
NM_001081087
|
Kbtbd10
|
kelch repeat and BTB (POZ) domain containing 10
|
|
chr5_-_17341713
|
5.450
|
NM_001159555
NM_007643
|
Cd36
|
CD36 antigen
|
|
chr2_-_76820599
|
5.390
|
NM_011652
NM_028004
|
Ttn
|
titin
|
|
chr8_-_25549374
|
5.373
|
|
1810011O10Rik
|
RIKEN cDNA 1810011O10 gene
|
|
chrX_+_154137114
|
5.358
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr1_+_45368443
|
5.316
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr19_-_5802596
|
5.308
|
|
Ramp2
Malat1
|
receptor (calcitonin) activity modifying protein 2
metastasis associated lung adenocarcinoma transcript 1 (non-coding RNA)
|
|
chr16_+_38362258
|
5.272
|
NM_001081984
NM_022318
|
Popdc2
|
popeye domain containing 2
|
|
chr3_-_10208480
|
5.267
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr11_-_51799215
|
5.251
|
|
Ube2b
|
ubiquitin-conjugating enzyme E2B, RAD6 homology (S. cerevisiae)
|
|
chr1_-_172018982
|
5.228
|
NM_022563
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr5_-_17341534
|
5.188
|
|
Cd36
|
CD36 antigen
|
|
chr9_-_91255831
|
5.185
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr1_+_155756310
|
5.110
|
|
Glul
|
glutamate-ammonia ligase (glutamine synthetase)
|
|
chr16_+_30065419
|
5.064
|
NM_008235
|
Hes1
|
hairy and enhancer of split 1 (Drosophila)
|
|
chr1_-_66981900
|
5.041
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chr11_-_5703677
|
5.031
|
|
Pgam2
|
phosphoglycerate mutase 2
|
|
chr5_+_102910409
|
5.029
|
NM_029270
NM_146161
|
Arhgap24
|
Rho GTPase activating protein 24
|
|
chr17_-_14831058
|
4.954
|
|
Thbs2
|
thrombospondin 2
|
|
chr10_+_38685309
|
4.938
|
NM_010681
|
Lama4
|
laminin, alpha 4
|
|
chr12_-_104594210
|
4.930
|
NM_023049
|
Asb2
|
ankyrin repeat and SOCS box-containing 2
|
|
chr17_-_14831095
|
4.847
|
|
Thbs2
|
thrombospondin 2
|
|
chr7_+_88202481
|
4.845
|
NM_054085
|
Alpk3
|
alpha-kinase 3
|
|
chr1_-_66991625
|
4.741
|
NM_021285
|
Myl1
|
myosin, light polypeptide 1
|
|
chr3_+_123058707
|
4.735
|
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae)
|
|
chr5_-_17394776
|
4.733
|
NM_001159558
|
Cd36
|
CD36 antigen
|
|
chr11_+_29642253
|
4.596
|
|
Rtn4
|
reticulon 4
|
|
chr7_-_116879475
|
4.587
|
|
Tmem9b
|
TMEM9 domain family, member B
|
|
chr3_+_19857053
|
4.581
|
NM_001042611
NM_007752
|
Cp
|
ceruloplasmin
|
|
chr11_+_29618531
|
4.565
|
NM_024226
|
Rtn4
|
reticulon 4
|
|
chr16_-_84955086
|
4.561
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr17_+_75784448
|
4.552
|
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr18_+_69843588
|
4.431
|
|
Tcf4
|
transcription factor 4
|
|
chr2_-_84490827
|
4.425
|
NM_001085448
NM_001085449
NM_001085450
NM_007615
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chr8_-_96496001
|
4.423
|
|
Amfr
|
autocrine motility factor receptor
|
|
chr2_+_138104219
|
4.418
|
NM_145534
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr10_-_36855292
|
4.417
|
|
Marcks
|
myristoylated alanine rich protein kinase C substrate
|
|
chr4_-_60083321
|
4.409
|
NM_001134675
|
Mup7
|
major urinary protein 7
|
|
chr2_+_20644000
|
4.403
|
|
Etl4
|
enhancer trap locus 4
|
|
chr10_+_38685424
|
4.384
|
|
Lama4
|
laminin, alpha 4
|
|
chr11_+_70471297
|
4.370
|
NM_007933
|
Eno3
|
enolase 3, beta muscle
|
|
chr5_+_122550987
|
4.345
|
NM_010861
|
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow
|
|
chr11_-_55208276
|
4.339
|
|
|
|
|
chr9_+_50373472
|
4.320
|
NM_008360
|
Il18
|
interleukin 18
|
|
chr12_+_73889998
|
4.309
|
|
Ppm1a
|
protein phosphatase 1A, magnesium dependent, alpha isoform
|
|
chr17_+_75835208
|
4.282
|
NM_001166493
|
Rasgrp3
|
RAS, guanyl releasing protein 3
|
|
chr17_-_14831233
|
4.271
|
|
Thbs2
|
thrombospondin 2
|
|
chr18_+_37663917
|
4.268
|
NM_053145
|
Pcdhb20
|
protocadherin beta 20
|
|
chr4_-_75779379
|
4.253
|
NM_001014288
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr8_-_64381465
|
4.250
|
NM_001081390
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr4_+_133871031
|
4.237
|
NM_001039048
|
Trim63
|
tripartite motif-containing 63
|
|
chr1_-_172040621
|
4.186
|
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr14_-_21483775
|
4.182
|
|
Synpo2l
|
synaptopodin 2-like
|
|
chrX_+_154137049
|
4.181
|
NM_025357
|
Smpx
|
small muscle protein, X-linked
|
|
chr16_+_32271553
|
4.148
|
NM_183283
|
2310010M20Rik
|
RIKEN cDNA 2310010M20 gene
|
|
chr1_-_174221511
|
4.106
|
|
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr8_+_131028275
|
4.102
|
|
Nrp1
|
neuropilin 1
|
|
chr17_-_7997838
|
4.072
|
NM_001081416
|
Fndc1
|
fibronectin type III domain containing 1
|
|
chr2_-_115890747
|
4.062
|
NM_001136072
NM_001159567
NM_001159568
NM_010825
|
Meis2
|
Meis homeobox 2
|
|
chr1_+_137732910
|
4.036
|
|
Tnnt2
|
troponin T2, cardiac
|
|
chr3_+_66023884
|
3.931
|
|
Ptx3
|
pentraxin related gene
|
|
chr17_-_14831262
|
3.927
|
NM_011581
|
Thbs2
|
thrombospondin 2
|