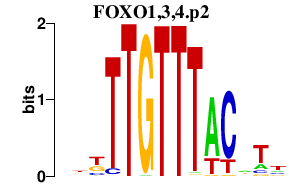
|
chr1_+_45390415
|
21.306
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_45368443
|
12.175
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_90031368
|
10.673
|
NM_145079
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A
|
|
chr4_+_104829867
|
10.643
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr15_+_3220766
|
9.512
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr15_+_3230053
|
9.475
|
|
|
|
|
chr15_+_3220994
|
9.414
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr15_+_3220999
|
9.333
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr3_-_10208480
|
9.088
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr4_+_104829963
|
8.898
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr4_+_104830022
|
8.748
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr4_+_104829951
|
8.746
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr6_+_15750966
|
8.592
|
|
Mdfic
|
MyoD family inhibitor domain containing
|
|
chr13_+_49703402
|
8.446
|
NM_008760
|
Ogn
|
osteoglycin
|
|
chr16_+_48872720
|
8.144
|
NM_181596
|
Retnlg
|
resistin like gamma
|
|
chr4_+_94405963
|
8.139
|
NM_013690
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr5_-_98459943
|
8.041
|
NM_133738
|
Antxr2
|
anthrax toxin receptor 2
|
|
chr15_+_102289600
|
7.998
|
NM_001170911
NM_025385
|
Prr13
|
proline rich 13
|
|
chr2_-_84614960
|
7.817
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr1_+_133867185
|
7.634
|
NM_145977
|
Slc45a3
|
solute carrier family 45, member 3
|
|
chr15_-_74828317
|
7.576
|
NM_010738
|
Ly6a
|
lymphocyte antigen 6 complex, locus A
|
|
chr1_+_89999782
|
7.465
|
NM_201410
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr4_+_94406190
|
7.346
|
|
Tek
|
endothelial-specific receptor tyrosine kinase
|
|
chr1_+_45368382
|
7.345
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr5_+_145888588
|
7.115
|
|
Arpc1b
|
actin related protein 2/3 complex, subunit 1B
|
|
chr10_+_87323855
|
6.838
|
NM_001111274
NM_001111276
NM_184052
|
Igf1
|
insulin-like growth factor 1
|
|
chr3_+_79690082
|
6.829
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr3_+_79689897
|
6.717
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr6_-_92851434
|
6.695
|
NM_175314
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9
|
|
chr13_+_49639847
|
6.375
|
|
Aspn
|
asporin
|
|
chr15_+_83393975
|
6.252
|
NM_009775
|
Tspo
|
translocator protein
|
|
chr6_-_3349415
|
5.757
|
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr15_-_37162895
|
5.695
|
|
Gm16136
|
predicted gene 16136
|
|
chr4_+_114761644
|
5.486
|
|
Pdzk1ip1
|
PDZK1 interacting protein 1
|
|
chr3_-_143868114
|
5.425
|
|
Lmo4
|
LIM domain only 4
|
|
chr16_+_35002066
|
5.342
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr1_-_72905107
|
5.271
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr6_+_34813131
|
5.225
|
NM_197986
|
Tmem140
|
transmembrane protein 140
|
|
chr6_+_72494474
|
5.197
|
|
Capg
|
capping protein (actin filament), gelsolin-like
|
|
chr11_-_120587426
|
5.154
|
|
|
|
|
chr9_-_21940154
|
5.077
|
NM_001102405
|
Acp5
|
acid phosphatase 5, tartrate resistant
|
|
chr6_-_5446277
|
5.071
|
NM_013743
|
Pdk4
|
pyruvate dehydrogenase kinase, isoenzyme 4
|
|
chr13_-_64468380
|
5.005
|
|
Ctsl
|
cathepsin L
|
|
chr7_-_26175113
|
4.985
|
NM_001039507
|
Lipe
|
lipase, hormone sensitive
|
|
chr3_+_54165028
|
4.927
|
NM_001198765
NM_001198766
NM_015784
|
Postn
|
periostin, osteoblast specific factor
|
|
chr17_+_34442837
|
4.902
|
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr19_+_12873063
|
4.892
|
NM_134152
|
Lpxn
|
leupaxin
|
|
chr5_+_115346942
|
4.892
|
NM_011854
|
Oasl2
|
2'-5' oligoadenylate synthetase-like 2
|
|
chr6_+_72494378
|
4.890
|
NM_007599
|
Capg
|
capping protein (actin filament), gelsolin-like
|
|
chr11_-_106576585
|
4.876
|
NM_001032378
NM_008816
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr6_-_101266541
|
4.874
|
|
9530086O07Rik
|
RIKEN cDNA 9530086O07 gene
|
|
chr19_+_29441927
|
4.871
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr12_-_27129497
|
4.860
|
|
Rsad2
|
radical S-adenosyl methionine domain containing 2
|
|
chr4_+_57907600
|
4.824
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr17_+_34400152
|
4.807
|
NM_207105
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1
|
|
chr9_+_108239210
|
4.797
|
|
Rhoa
|
ras homolog gene family, member A
|
|
chr13_-_74832039
|
4.777
|
|
Cast
|
calpastatin
|
|
chr17_+_34442808
|
4.711
|
NM_010382
|
H2-Eb1
|
histocompatibility 2, class II antigen E beta
|
|
chr2_-_52194317
|
4.706
|
NM_010889
|
Neb
|
nebulin
|
|
chr11_-_106576550
|
4.686
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr4_+_116265582
|
4.672
|
|
Gpbp1l1
|
GC-rich promoter binding protein 1-like 1
|
|
chr10_+_51200463
|
4.630
|
NM_008147
|
Gp49a
|
glycoprotein 49 A
|
|
chr7_-_3796619
|
4.620
|
|
Pira1
Pira2
Lilrb3
|
paired-Ig-like receptor A1
paired-Ig-like receptor A2
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
|
|
chr10_+_79457716
|
4.620
|
|
Cnn2
|
calponin 2
|
|
chr17_+_34341467
|
4.597
|
|
Tap2
|
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP)
|
|
chr11_+_100931369
|
4.585
|
NM_013792
|
Naglu
|
alpha-N-acetylglucosaminidase (Sanfilippo disease IIIB)
|
|
chr11_-_106576380
|
4.569
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr7_-_3849680
|
4.567
|
NM_001166672
NM_011088
NM_011091
NM_011094
|
Gm14548
Pira11
Pira4
Pira5
Pira7
Lilrb3
|
predicted gene 14548
paired-Ig-like receptor A11
paired-Ig-like receptor A4
paired-Ig-like receptor A5
paired-Ig-like receptor A7
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3
|
|
chr10_+_57514349
|
4.550
|
NM_020561
|
Smpdl3a
|
sphingomyelin phosphodiesterase, acid-like 3A
|
|
chr2_+_145647868
|
4.539
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr4_+_134479542
|
4.530
|
|
D4Wsu53e
|
DNA segment, Chr 4, Wayne State University 53, expressed
|
|
chr12_-_104681842
|
4.472
|
NM_029803
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A
|
|
chr7_+_4225744
|
4.465
|
NM_011093
NM_008848
|
Pira6
|
paired-Ig-like receptor A6
|
|
chr7_-_125629996
|
4.455
|
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr6_+_4490515
|
4.451
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr5_-_17341534
|
4.426
|
|
Cd36
|
CD36 antigen
|
|
chr9_+_50302624
|
4.417
|
NM_029639
|
1600029D21Rik
|
RIKEN cDNA 1600029D21 gene
|
|
chr6_-_3349470
|
4.383
|
NM_010156
|
Samd9l
|
sterile alpha motif domain containing 9-like
|
|
chr4_+_97444312
|
4.374
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr11_-_106576464
|
4.332
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr13_+_49704471
|
4.311
|
|
Ogn
|
osteoglycin
|
|
chr2_+_87366184
|
4.311
|
|
Srp9
|
signal recognition particle 9
|
|
chr15_+_101974616
|
4.234
|
NM_008344
|
Igfbp6
|
insulin-like growth factor binding protein 6
|
|
chr15_+_101974978
|
4.177
|
|
Igfbp6
|
insulin-like growth factor binding protein 6
|
|
chr3_+_137940628
|
4.137
|
|
Adh1
|
alcohol dehydrogenase 1 (class I)
|
|
chr2_+_103911166
|
4.117
|
NM_181858
|
Cd59b
|
CD59b antigen
|
|
chr7_+_148021503
|
4.110
|
|
1190003J15Rik
|
RIKEN cDNA 1190003J15 gene
|
|
chr2_-_32837527
|
4.093
|
NM_019488
|
Slc2a8
|
solute carrier family 2, (facilitated glucose transporter), member 8
|
|
chr9_-_115152895
|
4.070
|
|
Stt3b
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr4_-_19849637
|
4.056
|
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2
|
|
chr16_+_45093784
|
4.039
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr2_-_60589059
|
4.016
|
|
Rbms1
|
RNA binding motif, single stranded interacting protein 1
|
|
chr15_+_101974792
|
4.004
|
|
Igfbp6
|
insulin-like growth factor binding protein 6
|
|
chr5_-_17341512
|
4.000
|
|
Cd36
|
CD36 antigen
|
|
chr8_+_46970814
|
3.979
|
NM_016798
|
Pdlim3
|
PDZ and LIM domain 3
|
|
chr3_+_137940608
|
3.953
|
NM_007409
|
Adh1
|
alcohol dehydrogenase 1 (class I)
|
|
chr3_+_79689851
|
3.922
|
NM_133187
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr4_+_134479583
|
3.906
|
|
D4Wsu53e
|
DNA segment, Chr 4, Wayne State University 53, expressed
|
|
chr10_-_117252126
|
3.868
|
|
|
|
|
chr14_+_51716032
|
3.859
|
|
Ang
Rnase4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family 4
|
|
chr10_+_96942124
|
3.849
|
NM_001190451
|
Dcn
|
decorin
|
|
chr13_+_49678115
|
3.830
|
NM_012050
|
Omd
|
osteomodulin
|
|
chr17_+_24879324
|
3.827
|
|
Sepx1
|
selenoprotein X 1
|
|
chr16_+_43619057
|
3.827
|
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr2_+_122060526
|
3.815
|
NM_146126
|
Sord
|
sorbitol dehydrogenase
|
|
chr19_-_41288465
|
3.786
|
|
Tm9sf3
|
transmembrane 9 superfamily member 3
|
|
chr2_-_163470870
|
3.780
|
NM_012032
|
Serinc3
|
serine incorporator 3
|
|
chr6_-_122806042
|
3.766
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr9_-_21935019
|
3.714
|
NM_007388
|
Acp5
|
acid phosphatase 5, tartrate resistant
|
|
chr9_+_114373337
|
3.714
|
|
Glb1
|
galactosidase, beta 1
|
|
chr16_+_25802001
|
3.713
|
NM_001127262
NM_001127263
NM_001127264
NM_001127265
NM_011641
|
Trp63
|
transformation related protein 63
|
|
chr4_-_118162385
|
3.712
|
NM_011587
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr3_+_96361879
|
3.696
|
NM_001009935
NM_023719
|
Txnip
|
thioredoxin interacting protein
|
|
chr4_-_118162354
|
3.659
|
|
Tie1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr8_+_109127245
|
3.654
|
NM_009864
|
Cdh1
|
cadherin 1
|
|
chr10_+_87322774
|
3.641
|
|
Igf1
|
insulin-like growth factor 1
|
|
chr6_+_122463595
|
3.639
|
|
Mfap5
|
microfibrillar associated protein 5
|
|
chr17_+_29227908
|
3.618
|
NM_001111099
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21)
|
|
chr13_-_37142112
|
3.563
|
NM_001166391
|
F13a1
|
coagulation factor XIII, A1 subunit
|
|
chr5_+_7960430
|
3.561
|
NM_054098
|
Steap4
|
STEAP family member 4
|
|
chr15_-_76853069
|
3.550
|
NM_001164048
NM_001164047
|
Mb
|
myoglobin
|
|
chr2_-_66512442
|
3.511
|
|
Scn7a
|
sodium channel, voltage-gated, type VII, alpha
|
|
chr10_-_30338265
|
3.506
|
|
Hint3
|
histidine triad nucleotide binding protein 3
|
|
chr6_+_17231172
|
3.504
|
|
Cav2
|
caveolin 2
|
|
chr13_-_32873647
|
3.459
|
NM_001166030
|
Mylk4
|
myosin light chain kinase family, member 4
|
|
chr11_+_83517628
|
3.451
|
|
Gm11428
|
predicted gene 11428
|
|
chr19_+_3935169
|
3.401
|
NM_001161428
NM_019449
|
Unc93b1
|
unc-93 homolog B1 (C. elegans)
|
|
chr15_-_3329691
|
3.391
|
|
|
|
|
chr6_-_122806020
|
3.390
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr14_+_31692725
|
3.388
|
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr1_-_173582969
|
3.371
|
NM_144539
|
Slamf7
|
SLAM family member 7
|
|
chr7_-_3629111
|
3.371
|
NM_181820
|
Tmc4
|
transmembrane channel-like gene family 4
|
|
chr4_-_55545211
|
3.367
|
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr16_+_34995000
|
3.321
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr12_+_82049290
|
3.317
|
|
Srsf5
|
serine/arginine-rich splicing factor 5
|
|
chr5_+_43907004
|
3.303
|
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7
|
|
chr4_+_144482722
|
3.296
|
NM_001172424
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_+_137635977
|
3.285
|
|
Csrp1
|
cysteine and glycine-rich protein 1
|
|
chr7_-_87547589
|
3.276
|
NM_011046
|
Furin
|
furin (paired basic amino acid cleaving enzyme)
|
|
chr1_+_132762248
|
3.275
|
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|
|
chr10_-_58904445
|
3.251
|
NM_010959
|
Oit3
|
oncoprotein induced transcript 3
|
|
chr11_+_83517517
|
3.239
|
NM_001081957
|
Gm11428
|
predicted gene 11428
|
|
chr2_-_167515255
|
3.208
|
|
Tmem189
|
transmembrane protein 189
|
|
chr19_+_21354915
|
3.189
|
|
Zfand5
|
zinc finger, AN1-type domain 5
|
|
chr19_+_32670279
|
3.154
|
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2
|
|
chr3_+_65332306
|
3.150
|
NM_178892
|
Tiparp
|
TCDD-inducible poly(ADP-ribose) polymerase
|
|
chr10_-_61960613
|
3.133
|
|
Srgn
|
serglycin
|
|
chr7_-_133819870
|
3.132
|
NM_133670
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1
|
|
chr10_-_127613623
|
3.125
|
NM_019711
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr7_+_148641122
|
3.118
|
|
Pnpla2
|
patatin-like phospholipase domain containing 2
|
|
chr9_-_110959702
|
3.111
|
NM_017466
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2
|
|
chr4_-_133650937
|
3.103
|
NM_013706
|
Cd52
|
CD52 antigen
|
|
chr3_-_90318178
|
3.102
|
NM_011309
|
S100a1
|
S100 calcium binding protein A1
|
|
chr9_-_66881744
|
3.084
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr3_-_113277601
|
3.082
|
NM_007446
|
Amy1
|
amylase 1, salivary
|
|
chr10_-_127613494
|
3.079
|
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2
|
|
chr6_-_127076410
|
3.078
|
|
Ccnd2
|
cyclin D2
|
|
chr3_-_20174589
|
3.068
|
NM_029706
|
Cpb1
|
carboxypeptidase B1 (tissue)
|
|
chr19_+_12873115
|
3.068
|
|
Lpxn
|
leupaxin
|
|
chr10_-_83989221
|
3.058
|
|
Ckap4
|
cytoskeleton-associated protein 4
|
|
chr11_-_83388330
|
3.047
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr15_-_82184631
|
3.039
|
NM_025987
|
Ndufa6
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6 (B14)
|
|
chr19_+_5492584
|
3.030
|
|
|
|
|
chr14_+_120830603
|
3.016
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr5_-_130478687
|
2.994
|
NM_010368
|
Gusb
|
glucuronidase, beta
|
|
chr9_-_15105965
|
2.985
|
NM_133732
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr1_-_45559700
|
2.966
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr11_-_118181503
|
2.958
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr11_-_118162557
|
2.943
|
|
|
|
|
chr4_-_46579311
|
2.939
|
NM_178893
|
Coro2a
|
coronin, actin binding protein 2A
|
|
chr1_-_99873795
|
2.933
|
|
Pam
|
peptidylglycine alpha-amidating monooxygenase
|
|
chr2_-_163470826
|
2.920
|
|
Serinc3
|
serine incorporator 3
|
|
chrX_-_35793335
|
2.907
|
|
Lamp2
|
lysosomal-associated membrane protein 2
|
|
chr10_+_126857922
|
2.905
|
NM_001168293
NM_027900
|
R3hdm2
|
R3H domain containing 2
|
|
chr12_-_85791928
|
2.902
|
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1
|
|
chr2_-_163470787
|
2.901
|
|
Serinc3
|
serine incorporator 3
|
|
chr18_+_5046584
|
2.875
|
NM_153153
|
Svil
|
supervillin
|
|
chr8_-_72433344
|
2.873
|
|
Gatad2a
|
GATA zinc finger domain containing 2A
|
|
chr2_+_72244049
|
2.871
|
|
B230120H23Rik
|
RIKEN cDNA B230120H23 gene
|
|
chr4_+_57867433
|
2.818
|
NM_001035533
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr12_+_112783003
|
2.800
|
|
Eif5
|
eukaryotic translation initiation factor 5
|
|
chr1_-_75139207
|
2.800
|
|
1810031K17Rik
|
RIKEN cDNA 1810031K17 gene
|
|
chr4_-_60889249
|
2.793
|
NM_001134674
|
Mup13
Mup19
Mup1
|
major urinary protein 13
major urinary protein 19
major urinary protein 1
|
|
chr4_-_57156490
|
2.779
|
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr4_-_86315874
|
2.777
|
NM_007408
|
Plin2
|
perilipin 2
|
|
chr3_+_96049419
|
2.776
|
NM_013549
NM_178212
|
Hist2h3c1
Hist2h2aa1
Hist2h2aa2
|
histone cluster 2, H3c1
histone cluster 2, H2aa1
histone cluster 2, H2aa2
|
|
chr7_+_28902406
|
2.769
|
|
Fcgbp
|
Fc fragment of IgG binding protein
|
|
chr9_+_89094190
|
2.767
|
|
Bcl2a1a
|
B-cell leukemia/lymphoma 2 related protein A1a
|
|
chr16_+_45093816
|
2.752
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr3_-_116671869
|
2.751
|
NM_023245
|
Palmd
|
palmdelphin
|
|
chr6_+_115372083
|
2.742
|
NM_011146
|
Pparg
|
peroxisome proliferator activated receptor gamma
|
|
chr16_+_48872756
|
2.740
|
|
Retnlg
|
resistin like gamma
|
|
chr3_-_89084872
|
2.726
|
NM_001162425
|
Efna1
|
ephrin A1
|
|
chr17_-_56261311
|
2.702
|
NM_029796
|
Lrg1
|
leucine-rich alpha-2-glycoprotein 1
|
|
chr2_-_163462584
|
2.699
|
|
Serinc3
|
serine incorporator 3
|
|
chr6_-_16848352
|
2.689
|
NM_031198
|
Tcfec
|
transcription factor EC
|
|
chr1_+_16678513
|
2.688
|
|
Ly96
|
lymphocyte antigen 96
|
|
chr7_-_3820753
|
2.685
|
|
Gm10693
|
predicted pseudogene 10693
|
|
chr13_-_100972022
|
2.675
|
NM_001126182
NM_010872
|
Naip2
|
NLR family, apoptosis inhibitory protein 2
|
|
chr1_+_132762353
|
2.673
|
NM_026976
|
Faim3
|
Fas apoptotic inhibitory molecule 3
|