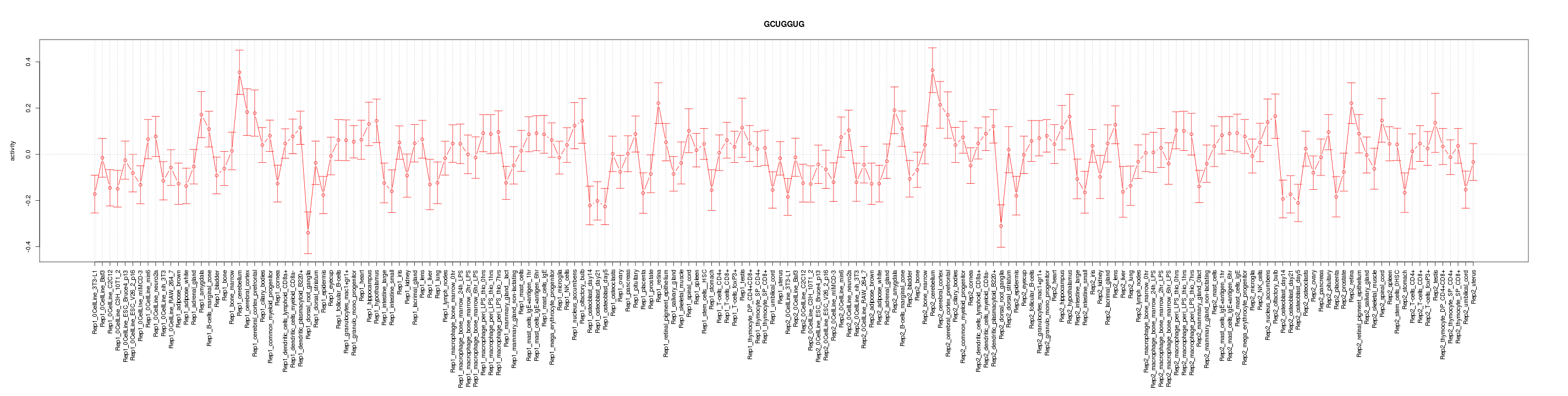
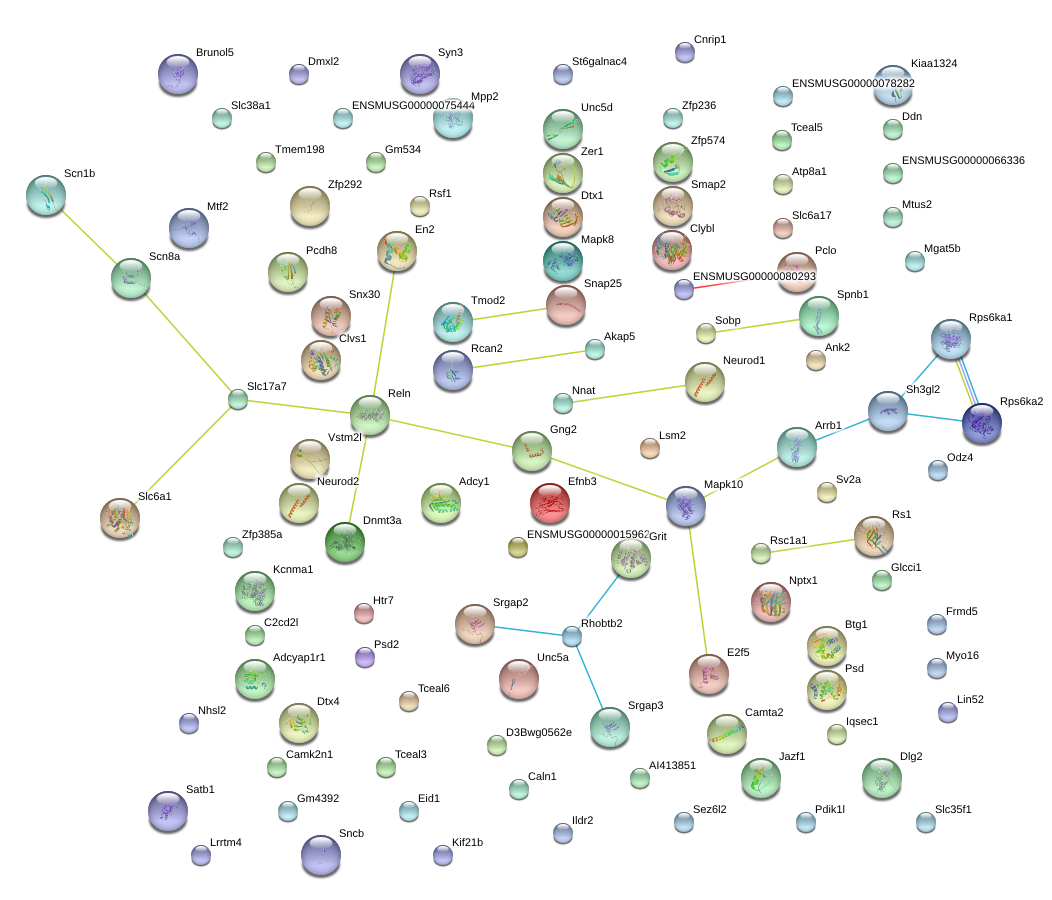
|
chr2_+_136539143
|
24.278
|
NM_011428
|
Snap25
|
synaptosomal-associated protein 25
|
|
chr5_-_121161656
|
21.361
|
NM_008052
|
Dtx1
|
deltex 1 homolog (Drosophila)
|
|
chr5_+_130845319
|
20.625
|
NM_181045
|
Caln1
|
calneuron 1
|
|
chr7_+_134094045
|
19.357
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr3_+_95985065
|
19.094
|
NM_022030
|
Sv2a
|
synaptic vesicle glycoprotein 2 a
|
|
chr5_+_130924661
|
18.803
|
NM_021371
|
Caln1
|
calneuron 1
|
|
chr4_+_84851330
|
16.206
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr18_+_36124483
|
15.142
|
NM_028707
|
Psd2
|
pleckstrin and Sec7 domain containing 2
|
|
chr10_-_80945445
|
14.848
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr6_+_114232618
|
14.620
|
NM_178703
|
Slc6a1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr12_+_85792371
|
14.231
|
NM_173756
|
Lin52
|
lin-52 homolog (C. elegans)
|
|
chr7_+_52419289
|
13.788
|
NM_182993
|
Slc17a7
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7
|
|
chr14_-_20796411
|
13.710
|
NM_001038637
NM_010315
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr15_+_100766701
|
12.837
|
NM_011323
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr8_-_30330079
|
12.406
|
NM_153135
|
Unc5d
|
unc-5 homolog D (C. elegans)
|
|
chr11_-_119409041
|
12.090
|
NM_008730
|
Nptx1
|
neuronal pentraxin 1
|
|
chr15_+_100701068
|
12.023
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr2_-_29979973
|
11.692
|
NM_178694
|
Zer1
|
zer-1 homolog (C. elegans)
|
|
chr2_+_157740281
|
11.055
|
NM_198627
|
Vstm2l
|
V-set and transmembrane domain containing 2-like
|
|
chr13_-_54867800
|
11.043
|
NM_033610
|
Sncb
|
synuclein, beta
|
|
chr2_-_79296732
|
10.687
|
NM_010894
|
Neurod1
|
neurogenic differentiation 1
|
|
chr4_+_138011061
|
10.567
|
NM_025451
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr4_+_9196439
|
10.486
|
NM_028940
|
Clvs1
|
clavesin 1
|
|
chr6_+_8459595
|
10.379
|
NM_133236
|
Glcci1
|
glucocorticoid induced transcript 1
|
|
chrX_-_131745187
|
10.144
|
NM_025355
|
Tceal6
|
transcription elongation factor A (SII)-like 6
|
|
chr10_+_52410306
|
10.070
|
NM_178675
|
Slc35f1
|
solute carrier family 35, member F1
|
|
chr1_+_75476106
|
10.006
|
NM_177056
|
Tmem198
|
transmembrane protein 198
|
|
chr6_+_55401971
|
9.857
|
NM_001025372
NM_007407
|
Adcyap1r1
|
adenylate cyclase activating polypeptide 1 receptor 1
|
|
chr5_-_21850490
|
9.743
|
NM_011261
|
Reln
|
reelin
|
|
chrX_-_132738383
|
9.589
|
NM_177919
|
Tceal5
|
transcription elongation factor A (SII)-like 5
|
|
chr2_-_121632749
|
9.504
|
NM_172673
|
Frmd5
|
FERM domain containing 5
|
|
chr15_-_96472392
|
9.450
|
NM_134086
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr11_-_69373707
|
9.120
|
NM_007911
|
Efnb3
|
ephrin B3
|
|
chr3_-_126646303
|
9.079
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr11_+_16951911
|
8.820
|
NM_029861
|
Cnrip1
|
cannabinoid receptor interacting protein 1
|
|
chr15_-_96473193
|
8.733
|
NM_001166458
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr1_+_138027943
|
8.700
|
NM_001039472
|
Kif21b
|
kinesin family member 21B
|
|
chr6_-_112897259
|
8.688
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr19_-_46401618
|
8.496
|
NM_028627
|
Psd
|
pleckstrin and Sec7 domain containing
|
|
chr2_+_32442949
|
8.237
|
NM_011373
|
St6galnac4
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 4
|
|
chr12_+_77425836
|
8.083
|
NM_001101471
|
Akap5
|
A kinase (PRKA) anchor protein 5
|
|
chr4_-_120689813
|
8.047
|
NM_133716
|
Smap2
|
stromal membrane-associated GTPase-activating protein 2
|
|
chr5_+_108494692
|
7.993
|
NM_013827
|
Mtf2
|
metal response element binding transcription factor 2
|
|
chr5_+_148768875
|
7.938
|
NM_029920
|
Mtus2
|
microtubule associated tumor suppressor candidate 2
|
|
chr9_-_54349409
|
7.770
|
NM_172771
|
Dmxl2
|
Dmx-like 2
|
|
chr17_+_44090139
|
7.717
|
NM_030598
|
Rcan2
|
regulator of calcineurin 2
|
|
chr6_-_52859613
|
7.659
|
NM_001168277
|
Jazf1
|
JAZF zinc finger 1
|
|
chr15_-_103170514
|
7.615
|
NM_013866
|
Zfp385a
|
zinc finger protein 385A
|
|
chrX_+_133200913
|
7.382
|
NM_001029978
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr5_-_103640272
|
7.102
|
NM_001081567
NM_009158
|
Mapk10
|
mitogen-activated protein kinase 10
|
|
chr14_-_70205351
|
6.877
|
NM_153514
|
Rhobtb2
|
Rho-related BTB domain containing 2
|
|
chr3_-_107320881
|
6.725
|
NM_172271
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17
|
|
chr9_-_75446888
|
6.613
|
NM_001038710
|
Tmod2
|
tropomodulin 2
|
|
chr19_-_12576485
|
6.578
|
NM_172442
|
Dtx4
|
deltex 4 homolog (Drosophila)
|
|
chr9_+_32031833
|
6.304
|
NM_177379
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr6_-_90666474
|
6.288
|
NM_001134383
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr7_-_31911884
|
6.270
|
NM_011322
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr5_+_28492235
|
6.145
|
NM_010134
|
En2
|
engrailed 2
|
|
chr12_-_77811533
|
5.965
|
NM_013675
|
Spnb1
|
spectrin beta 1
|
|
chr17_+_43938789
|
5.929
|
NM_207649
|
Rcan2
|
regulator of calcineurin 2
|
|
chr11_+_6963421
|
5.883
|
NM_009622
|
Adcy1
|
adenylate cyclase 1
|
|
chr11_+_116780161
|
5.796
|
NM_172948
|
Mgat5b
|
mannoside acetylglucosaminyltransferase 5, isoenzyme B
|
|
chr7_+_98239230
|
5.737
|
NM_011807
|
Dlg2
|
discs, large homolog 2 (Drosophila)
|
|
chr5_+_14514906
|
5.737
|
NM_001110796
NM_011995
|
Pclo
|
piccolo (presynaptic cytomatrix protein)
|
|
chr3_-_117063793
|
5.717
|
NM_177664
|
D3Bwg0562e
|
DNA segment, Chr 3, Brigham & Women's Genetics 0562 expressed
|
|
chr4_+_59818521
|
5.704
|
NM_172468
|
Snx30
|
sorting nexin family member 30
|
|
chr17_-_51965912
|
5.698
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr6_+_79969636
|
5.596
|
NM_178731
|
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4
|
|
chr6_-_90760116
|
5.585
|
NM_001134384
|
Iqsec1
|
IQ motif and Sec7 domain 1
|
|
chr9_-_75459125
|
5.550
|
NM_016711
|
Tmod2
|
tropomodulin 2
|
|
chr10_-_85961630
|
5.518
|
NM_001164495
NM_013722
|
Syn3
|
synapsin III
|
|
chrX_+_99044723
|
5.471
|
NM_001163610
|
Nhsl2
|
NHS-like 2
|
|
chr3_-_126701367
|
5.336
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr13_+_55050729
|
5.301
|
NM_153131
|
Unc5a
|
unc-5 homolog A (C. elegans)
|
|
chr11_-_70501586
|
5.258
|
NM_001190376
NM_001190378
NM_001190379
NM_178116
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr14_-_24823370
|
5.248
|
NM_010610
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1
|
|
chr2_+_125498825
|
5.246
|
NM_025613
|
Eid1
|
EP300 interacting inhibitor of differentiation 1
|
|
chr15_-_98638348
|
5.234
|
NM_001013741
|
Ddn
|
dendrin
|
|
chr10_-_42894259
|
5.153
|
NM_175407
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr5_-_68238654
|
5.069
|
NM_001038999
NM_009727
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr12_+_3806955
|
5.025
|
NM_007872
|
Dnmt3a
|
DNA methyltransferase 3A
|
|
chr7_+_104728398
|
4.962
|
NM_001081267
|
Rsf1
|
remodeling and spacing factor 1
|
|
chr9_-_44128228
|
4.952
|
NM_027909
|
C2cd2l
|
C2 calcium-dependent domain containing 2-like
|
|
chr4_-_133443662
|
4.949
|
NM_009097
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1
|
|
chr10_+_96079634
|
4.906
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr2_+_157385835
|
4.885
|
NM_010923
NM_180960
|
Nnat
|
neuronatin
|
|
chr7_+_103359146
|
4.854
|
NM_011858
|
Odz4
|
odd Oz/ten-m homolog 4 (Drosophila)
|
|
chr4_-_133843746
|
4.851
|
NM_146156
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chr4_-_34830178
|
4.784
|
NM_013889
|
Zfp292
|
zinc finger protein 292
|
|
chr7_+_106683983
|
4.742
|
NM_177231
NM_178220
|
Arrb1
|
arrestin, beta 1
|
|
chr17_-_51972514
|
4.732
|
NM_001163632
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr1_+_168184269
|
4.716
|
NM_001164528
|
Ildr2
|
immunoglobulin-like domain containing receptor 2
|
|
chr4_-_133843139
|
4.629
|
NM_001163794
|
Pdik1l
|
PDLIM1 interacting kinase 1 like
|
|
chrX_+_157205944
|
4.603
|
NM_011302
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr13_+_44826121
|
4.597
|
NM_021878
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr17_-_51951378
|
4.555
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr6_+_120314116
|
4.543
|
NM_145997
|
Kdm5a
|
lysine (K)-specific demethylase 5A
|
|
chr8_+_85387234
|
4.457
|
NM_177378
|
Rnf150
|
ring finger protein 150
|
|
chrX_+_165233030
|
4.451
|
NM_009707
|
Arhgap6
|
Rho GTPase activating protein 6
|
|
chr10_-_61802889
|
4.427
|
NM_001146100
|
Hk1
|
hexokinase 1
|
|
chr4_+_128561383
|
4.408
|
NM_178110
|
Trim62
|
tripartite motif-containing 62
|
|
chr2_-_121176719
|
4.364
|
NM_178795
|
Ppip5k1
|
diphosphoinositol pentakisphosphate kinase 1
|
|
chr8_+_90723069
|
4.337
|
NM_001164497
NM_001164498
NM_001164499
|
Papd5
|
PAP associated domain containing 5
|
|
chr15_-_84818368
|
4.289
|
NM_001033273
|
5031439G07Rik
|
RIKEN cDNA 5031439G07 gene
|
|
chr5_+_22252006
|
4.268
|
NM_001081231
NM_029990
|
Lhfpl3
|
lipoma HMGIC fusion partner-like 3
|
|
chr14_-_13178341
|
4.248
|
NM_080433
|
Fezf2
|
Fez family zinc finger 2
|
|
chr2_-_130468507
|
4.125
|
NM_197945
|
Prosapip1
|
ProSAPiP1 protein
|
|
chr2_+_143371807
|
4.122
|
NM_008792
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2
|
|
chr9_+_47338431
|
4.118
|
NM_001025600
NM_018770
NM_207675
NM_207676
|
Cadm1
|
cell adhesion molecule 1
|
|
chr13_+_118009379
|
4.089
|
NM_010330
|
Emb
|
embigin
|
|
chr8_-_70342105
|
4.085
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr1_+_194014677
|
3.953
|
NM_001038607
NM_010600
|
Kcnh1
|
potassium voltage-gated channel, subfamily H (eag-related), member 1
|
|
chr15_-_75397243
|
3.935
|
NM_001135688
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr14_-_94287950
|
3.929
|
NM_001081377
|
Pcdh9
|
protocadherin 9
|
|
chr4_-_132768403
|
3.905
|
NM_008154
|
Gpr3
|
G-protein coupled receptor 3
|
|
chr15_-_75397601
|
3.880
|
NM_011837
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr12_-_86311718
|
3.816
|
NM_178065
|
1110018G07Rik
|
RIKEN cDNA 1110018G07 gene
|
|
chr10_-_79017835
|
3.810
|
NM_027422
|
Mier2
|
mesoderm induction early response 1, family member 2
|
|
chr2_+_5872500
|
3.779
|
NM_001081132
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast)
|
|
chr2_-_39046224
|
3.748
|
NM_178778
|
Scai
|
suppressor of cancer cell invasion
|
|
chrX_-_34529439
|
3.707
|
NM_001177323
|
Sept6
|
septin 6
|
|
chr17_-_51971946
|
3.705
|
NM_009122
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr9_+_108728650
|
3.685
|
NM_080437
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr10_-_61842599
|
3.649
|
NM_010438
|
Hk1
|
hexokinase 1
|
|
chr10_-_9620837
|
3.614
|
NM_001081344
|
Stxbp5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr6_-_53018566
|
3.559
|
NM_173406
|
Jazf1
|
JAZF zinc finger 1
|
|
chr11_-_69227176
|
3.554
|
NM_001017426
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B
|
|
chr2_+_92061718
|
3.550
|
NM_001109690
|
Phf21a
|
PHD finger protein 21A
|
|
chr15_-_75397030
|
3.539
|
NM_001135689
|
Ly6h
|
lymphocyte antigen 6 complex, locus H
|
|
chr2_+_121782185
|
3.489
|
NM_212450
|
Ctdspl2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2
|
|
chr8_-_61390372
|
3.431
|
NM_175032
|
Galntl6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6
|
|
chr8_+_86423968
|
3.418
|
NM_181039
|
Lphn1
|
latrophilin 1
|
|
chr12_+_87762593
|
3.413
|
NM_011934
|
Esrrb
|
estrogen related receptor, beta
|
|
chr14_+_35633312
|
3.408
|
NM_008166
|
Grid1
|
glutamate receptor, ionotropic, delta 1
|
|
chr5_+_130007103
|
3.393
|
NM_172276
|
Sfswap
|
splicing factor, suppressor of white-apricot homolog (Drosophila)
|
|
chr17_-_49703661
|
3.361
|
NM_001008231
|
Daam2
|
dishevelled associated activator of morphogenesis 2
|
|
chr12_+_87582805
|
3.344
|
NM_027405
|
1700020O03Rik
|
RIKEN cDNA 1700020O03 gene
|
|
chr7_+_142729505
|
3.340
|
NM_011212
|
Ptpre
|
protein tyrosine phosphatase, receptor type, E
|
|
chrX_+_33868347
|
3.300
|
NM_028894
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3
|
|
chr2_+_92025639
|
3.258
|
NM_001109691
|
Phf21a
|
PHD finger protein 21A
|
|
chr15_-_96472600
|
3.206
|
NM_001166456
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr2_+_24909898
|
3.204
|
NM_001039386
NM_001039387
NM_001177654
NM_001177655
NM_020276
|
Nelf
|
nasal embryonic LHRH factor
|
|
chr6_-_48036561
|
3.129
|
NM_001163475
|
Zfp746
|
zinc finger protein 746
|
|
chr3_-_55987612
|
3.107
|
NM_030595
|
Nbea
|
neurobeachin
|
|
chr11_+_77306825
|
3.061
|
NM_001004144
|
Git1
|
G protein-coupled receptor kinase-interactor 1
|
|
chr5_-_121641282
|
3.046
|
NM_001109992
NM_011202
|
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11
|
|
chr14_+_28051224
|
3.045
|
NM_027871
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3
|
|
chr4_+_49072327
|
3.028
|
NM_178756
|
E130309F12Rik
|
RIKEN cDNA E130309F12 gene
|
|
chr11_+_93747419
|
3.008
|
NM_134012
|
Mbtd1
|
mbt domain containing 1
|
|
chr8_-_122364444
|
3.001
|
NM_028071
|
Cotl1
|
coactosin-like 1 (Dictyostelium)
|
|
chr15_+_100592177
|
2.988
|
NM_021530
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8
|
|
chr8_-_112692071
|
2.973
|
NM_007586
|
Calb2
|
calbindin 2
|
|
chr12_+_87702066
|
2.865
|
NM_001159500
|
Esrrb
|
estrogen related receptor, beta
|
|
chr5_-_121227617
|
2.774
|
NM_145226
|
Oas3
|
2'-5' oligoadenylate synthetase 3
|
|
chr12_-_47919695
|
2.766
|
NM_021361
|
Nova1
|
neuro-oncological ventral antigen 1
|
|
chr6_-_122287014
|
2.745
|
NM_001042623
NM_007905
|
Phc1
|
polyhomeotic-like 1 (Drosophila)
|
|
chr9_+_31923720
|
2.728
|
NM_001195632
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr11_+_69579376
|
2.715
|
NM_029348
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr18_+_78135191
|
2.693
|
NM_001195633
|
5430411K18Rik
|
RIKEN cDNA 5430411K18 gene
|
|
chr3_+_28162135
|
2.571
|
NM_001163007
NM_001163008
NM_001163009
NM_026910
|
Tnik
|
TRAF2 and NCK interacting kinase
|
|
chr2_+_14976877
|
2.566
|
NM_029466
|
Arl5b
|
ADP-ribosylation factor-like 5B
|
|
chr2_+_163899842
|
2.511
|
NM_021420
|
Stk4
|
serine/threonine kinase 4
|
|
chr5_+_73872293
|
2.478
|
NM_001190734
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr6_-_113145321
|
2.444
|
NM_177763
|
Lhfpl4
|
lipoma HMGIC fusion partner-like protein 4
|
|
chr2_+_4322020
|
2.403
|
NM_001177843
|
Frmd4a
|
FERM domain containing 4A
|
|
chr7_-_125386862
|
2.371
|
NM_001031814
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chr18_-_46685557
|
2.343
|
NM_173423
|
Fem1c
|
fem-1 homolog c (C.elegans)
|
|
chr1_+_75371871
|
2.334
|
NM_007463
|
Speg
|
SPEG complex locus
|
|
chr2_+_90744878
|
2.304
|
NM_025991
|
Kbtbd4
|
kelch repeat and BTB (POZ) domain containing 4
|
|
chr5_+_73882264
|
2.289
|
NM_001190733
NM_178896
|
Dcun1d4
|
DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae)
|
|
chr8_-_70434808
|
2.278
|
NM_030263
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr8_+_24168730
|
2.252
|
NM_031158
|
Ank1
|
ankyrin 1, erythroid
|
|
chr13_+_43880475
|
2.230
|
NM_009856
|
Cd83
|
CD83 antigen
|
|
chrX_+_148667763
|
2.180
|
NM_013668
|
Kdm5c
|
lysine (K)-specific demethylase 5C
|
|
chr14_-_70132309
|
2.120
|
NM_134078
|
Chmp7
|
CHMP family, member 7
|
|
chr11_+_102622745
|
2.111
|
NM_001110778
NM_009613
|
Adam11
|
a disintegrin and metallopeptidase domain 11
|
|
chr9_+_40076800
|
2.105
|
NM_001083917
NM_153522
NM_178227
|
Scn3b
|
sodium channel, voltage-gated, type III, beta
|
|
chr15_-_78548542
|
2.097
|
NM_183141
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2
|
|
chr17_-_25707599
|
2.088
|
NM_011447
|
Sox8
|
SRY-box containing gene 8
|
|
chr2_+_4073908
|
2.075
|
NM_172475
|
Frmd4a
|
FERM domain containing 4A
|
|
chr2_+_173563060
|
2.047
|
NM_019806
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C
|
|
chr1_+_193149772
|
2.030
|
NM_025864
|
Tmem206
|
transmembrane protein 206
|
|
chr1_+_129765349
|
2.015
|
NM_178690
|
Rab3gap1
|
RAB3 GTPase activating protein subunit 1
|
|
chr5_-_135164426
|
2.005
|
NM_010717
|
Limk1
|
LIM-domain containing, protein kinase
|
|
chr2_+_4480799
|
1.971
|
NM_001177844
|
Frmd4a
|
FERM domain containing 4A
|
|
chr2_-_38142866
|
1.958
|
NM_146122
|
Dennd1a
|
DENN/MADD domain containing 1A
|
|
chr11_-_71949346
|
1.953
|
NM_001024927
NM_001081641
|
Pitpnm3
|
PITPNM family member 3
|
|
chr17_+_27993426
|
1.916
|
NM_001080769
|
Uhrf1bp1
|
UHRF1 (ICBP90) binding protein 1
|
|
chr14_+_31528034
|
1.915
|
NM_019460
|
Sfmbt1
|
Scm-like with four mbt domains 1
|
|
chr1_-_136131981
|
1.906
|
NM_001008533
NM_001039510
|
Adora1
|
adenosine A1 receptor
|
|
chr5_+_122659745
|
1.877
|
NM_001042489
|
Hvcn1
|
hydrogen voltage-gated channel 1
|
|
chr12_-_4484059
|
1.862
|
NM_010881
|
Ncoa1
|
nuclear receptor coactivator 1
|
|
chr1_-_90598670
|
1.845
|
NM_177305
|
Arl4c
|
ADP-ribosylation factor-like 4C
|
|
chr17_+_15190022
|
1.819
|
NM_001034891
|
2210404J11Rik
|
RIKEN cDNA 2210404J11 gene
|
|
chr4_+_130348406
|
1.781
|
NM_011520
|
Sdc3
|
syndecan 3
|
|
chr2_+_154262114
|
1.761
|
NM_009823
NM_172860
|
Cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 2 (human)
|
|
chr8_-_70498472
|
1.760
|
NM_177698
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr5_+_122660225
|
1.758
|
NM_028752
|
Hvcn1
|
hydrogen voltage-gated channel 1
|
|
chr14_+_52730577
|
1.742
|
NM_001168515
NM_023879
|
Rpgrip1
|
retinitis pigmentosa GTPase regulator interacting protein 1
|
|
chr1_-_88567078
|
1.735
|
NM_010933
|
Nppc
|
natriuretic peptide type C
|