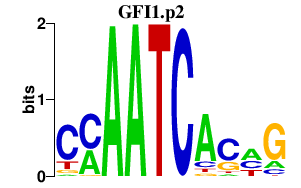
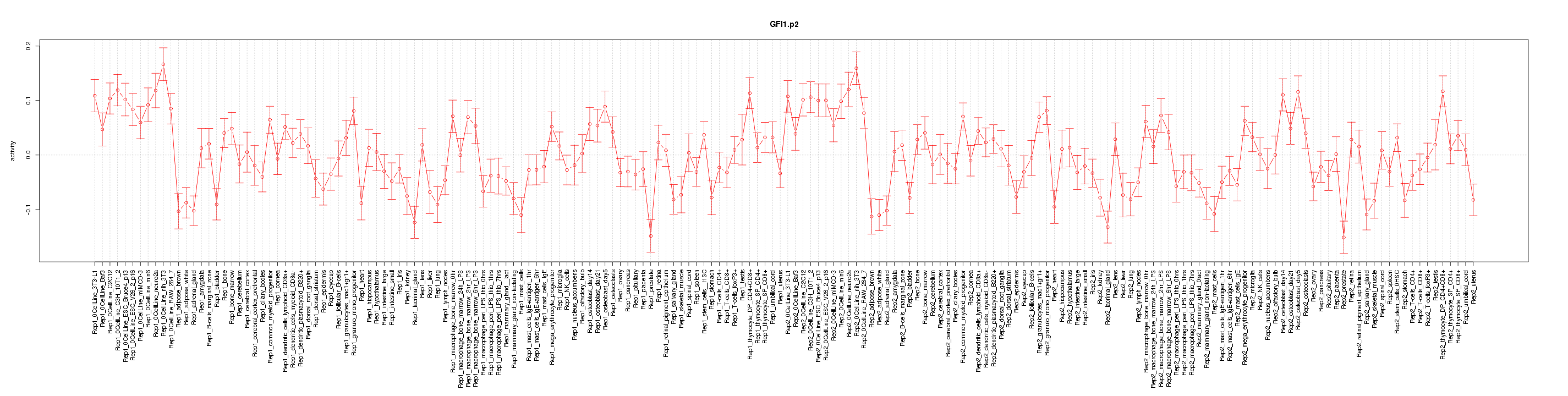
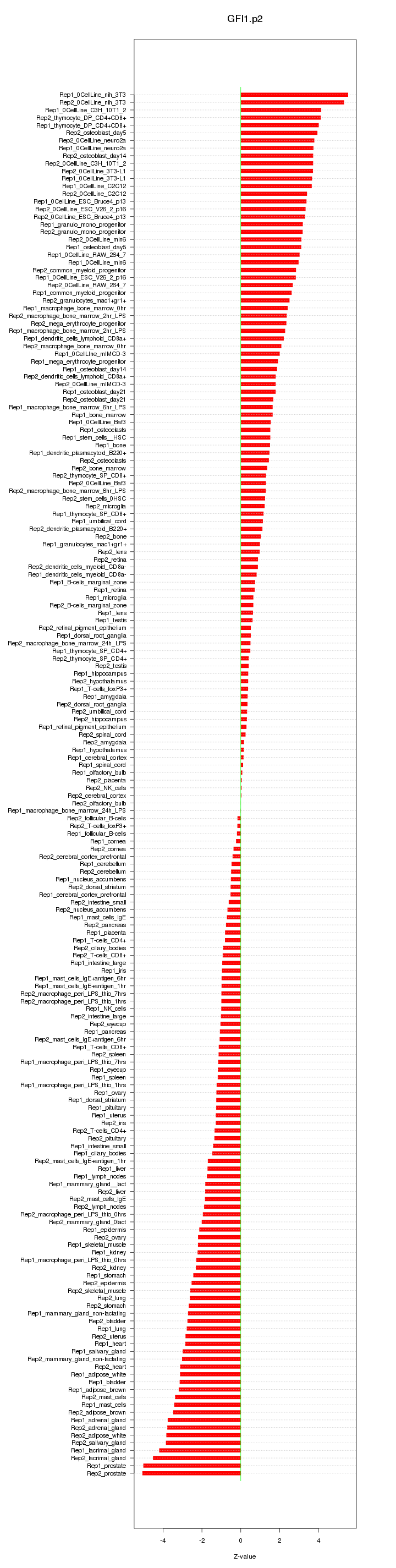
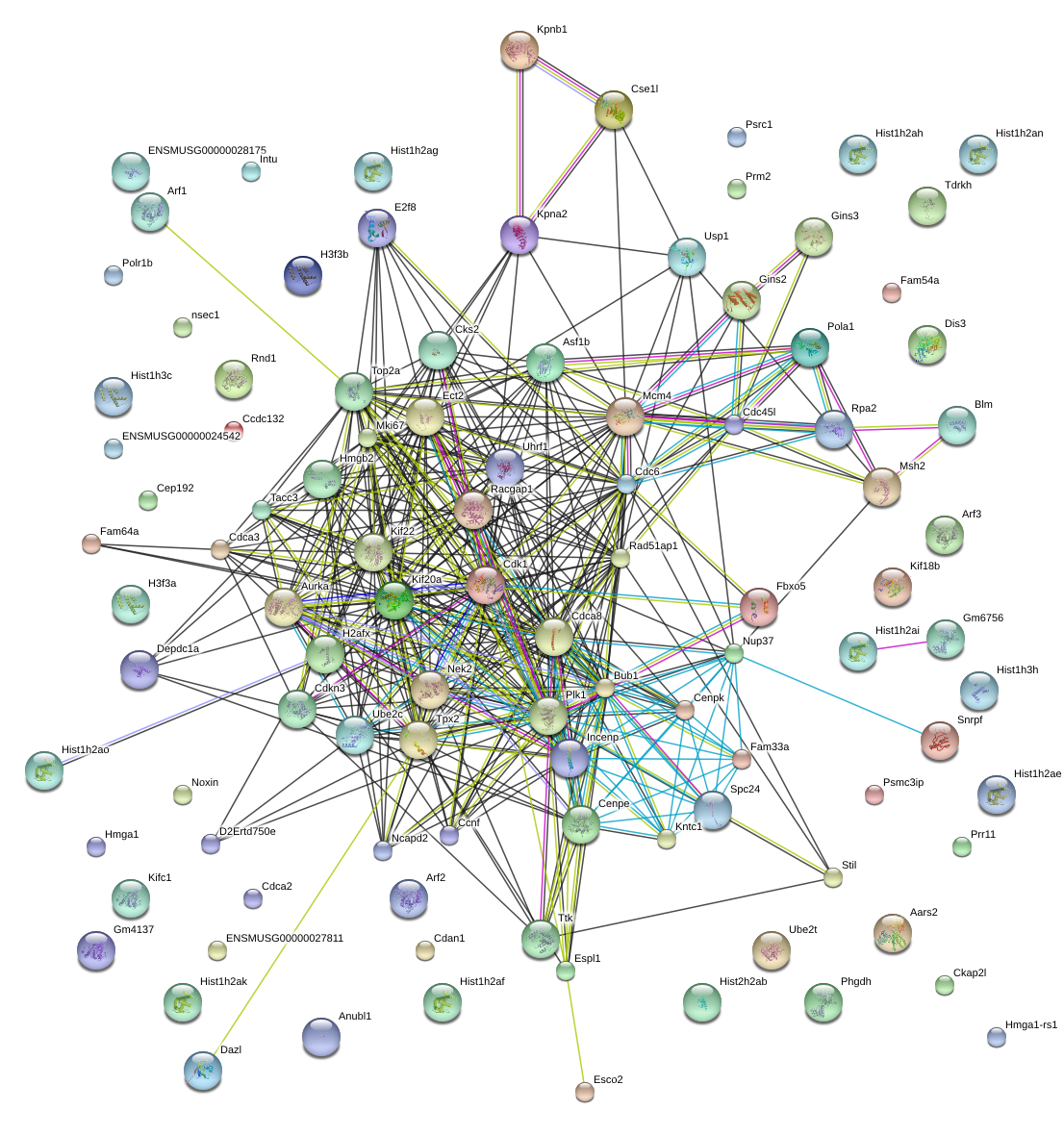
|
chr8_+_86479405
|
71.292
|
NM_024184
|
Asf1b
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr3_-_27052733
|
52.685
|
NM_001177625
NM_007900
|
Ect2
|
ect2 oncogene
|
|
chr14_-_68333666
|
50.852
|
NM_001110162
|
Cdca2
|
cell division cycle associated 2
|
|
chr3_-_27052794
|
45.522
|
NM_001177626
|
Ect2
|
ect2 oncogene
|
|
chr11_-_98885428
|
41.405
|
NM_011623
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr14_-_68333897
|
41.092
|
NM_175384
|
Cdca2
|
cell division cycle associated 2
|
|
chr2_+_164595415
|
36.748
|
NM_026785
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr16_-_18811669
|
36.287
|
NM_001161623
|
Cdc45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr2_+_164595447
|
36.161
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr2_-_172195998
|
35.553
|
NM_011497
|
Aurka
|
aurora kinase A
|
|
chr2_+_164595389
|
35.147
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr4_-_124614072
|
31.802
|
NM_026560
|
Cdca8
|
cell division cycle associated 8
|
|
chr13_+_51740600
|
29.980
|
NM_025415
|
Cks2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr13_+_51740804
|
29.448
|
|
Cks2
|
CDC28 protein kinase regulatory subunit 2
|
|
chr11_+_86922732
|
26.792
|
NM_025377
|
Fam33a
|
family with sequence similarity 33, member A
|
|
chr13_+_105019053
|
25.543
|
|
Cenpk
|
centromere protein K
|
|
chr17_-_34027575
|
25.005
|
NM_001195298
|
Kifc1
|
kinesin family member C1
|
|
chr6_-_125141525
|
25.002
|
NM_146171
|
Ncapd2
|
non-SMC condensin I complex, subunit D2
|
|
chr13_+_105019025
|
24.810
|
NM_181061
|
Cenpk
|
centromere protein K
|
|
chr16_-_15637390
|
23.861
|
NM_008565
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae)
|
|
chr17_-_34027488
|
23.279
|
|
Kifc1
|
kinesin family member C1
|
|
chr17_-_34027547
|
22.903
|
|
Kifc1
|
kinesin family member C1
|
|
chr3_+_159158396
|
20.627
|
NM_001172092
NM_001172093
NM_029523
|
Depdc1a
|
DEP domain containing 1a
|
|
chr5_+_124199713
|
20.441
|
NM_001042421
|
Kntc1
|
kinetochore associated 1
|
|
chr18_+_34784277
|
18.279
|
NM_001166407
|
Kif20a
|
kinesin family member 20A
|
|
chr15_-_99481964
|
18.094
|
NM_012025
|
Racgap1
|
Rac GTPase-activating protein 1
|
|
chr18_+_34784589
|
18.007
|
NM_009004
NM_001166406
|
Kif20a
|
kinesin family member 20A
|
|
chr8_+_59990639
|
17.539
|
NM_008252
|
Hmgb2
|
high mobility group box 2
|
|
chr10_-_93052348
|
17.485
|
NM_027246
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr16_-_18812064
|
17.035
|
NM_009862
|
Cdc45
|
cell division cycle 45 homolog (S. cerevisiae)
|
|
chr7_-_142908061
|
16.997
|
NM_001081117
|
Mki67
|
antigen identified by monoclonal antibody Ki 67
|
|
chr7_-_56136965
|
16.612
|
|
E2f8
|
E2F transcription factor 8
|
|
chr14_+_47380287
|
16.402
|
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr5_+_34001199
|
16.146
|
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr18_+_67959756
|
16.124
|
NM_027556
|
Cep192
|
centrosomal protein 192
|
|
chr11_-_86922163
|
15.992
|
NM_175563
|
Prr11
|
proline rich 11
|
|
chr13_+_105019467
|
15.706
|
NM_021790
|
Cenpk
|
centromere protein K
|
|
chr9_+_83728331
|
14.864
|
|
Ttk
|
Ttk protein kinase
|
|
chr9_+_83728295
|
14.599
|
NM_001110265
NM_009445
|
Ttk
|
Ttk protein kinase
|
|
chr10_-_68815606
|
14.517
|
NM_007659
|
Cdk1
|
cyclin-dependent kinase 1
|
|
chr9_-_21564742
|
14.200
|
|
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr14_+_47380215
|
14.056
|
NM_028222
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr11_-_106860750
|
14.019
|
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr7_+_129302998
|
13.857
|
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr1_+_193645351
|
13.849
|
NM_010892
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr15_+_102126686
|
13.779
|
NM_001014976
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr15_+_102126735
|
13.765
|
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae)
|
|
chr13_+_21902235
|
13.549
|
NM_178185
NM_001177544
|
Hist1h2ah
Hist1h2ap
Hist1h2ai
Hist1h2ao
|
histone cluster 1, H2ah
histone cluster 1, H2ap
histone cluster 1, H2ai
histone cluster 1, H2ao
|
|
chr19_-_9973901
|
13.274
|
NM_016692
|
Incenp
|
inner centromere protein
|
|
chr7_-_134185894
|
13.269
|
|
Kif22
|
kinesin family member 22
|
|
chr11_-_115885714
|
13.238
|
NM_008211
|
H3f3b
|
H3 histone, family 3B
|
|
chr7_-_134185928
|
13.192
|
NM_145588
|
Kif22
|
kinesin family member 22
|
|
chr10_+_4541138
|
13.142
|
|
Fbxo5
|
F-box protein 5
|
|
chr8_-_123112925
|
12.956
|
|
Gins2
|
GINS complex subunit 2 (Psf2 homolog)
|
|
chr3_+_94217197
|
12.905
|
NM_028307
|
Tdrkh
|
tudor and KH domain containing protein
|
|
chr5_+_34000745
|
12.857
|
NM_001040435
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr7_-_100022714
|
12.807
|
NM_001080995
|
4632434I11Rik
|
RIKEN cDNA 4632434I11 gene
|
|
chr13_-_21925343
|
12.650
|
NM_178185
NM_001177544
|
Hist1h2ah
Hist1h2ap
Hist1h2ai
Hist1h2ao
|
histone cluster 1, H2ah
histone cluster 1, H2ap
histone cluster 1, H2ai
histone cluster 1, H2ao
|
|
chr11_+_71855939
|
12.620
|
NM_144526
|
Fam64a
|
family with sequence similarity 64, member A
|
|
chr7_+_129303029
|
12.541
|
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr13_-_23663006
|
12.536
|
NM_178187
|
Hist1h2ae
|
histone cluster 1, H2ae
|
|
chr11_-_102786433
|
12.482
|
NM_197959
|
Kif18b
|
kinesin family member 18B
|
|
chr13_-_21845695
|
12.342
|
NM_178183
|
Hist1h2ak
|
histone cluster 1, H2ak
|
|
chr10_+_4541075
|
12.341
|
NM_025995
|
Fbxo5
|
F-box protein 5
|
|
chr11_-_97048933
|
12.295
|
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chr1_+_136859153
|
12.198
|
NM_026024
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative)
|
|
chr11_-_102786403
|
12.048
|
|
Kif18b
|
kinesin family member 18B
|
|
chr17_-_24388262
|
11.995
|
NM_007634
|
Ccnf
|
cyclin F
|
|
chr9_+_44142772
|
11.977
|
NM_010436
|
H2afx
|
H2A histone family, member X
|
|
chr11_+_98769162
|
11.892
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_103828032
|
11.802
|
|
Arf2
|
ADP-ribosylation factor 2
|
|
chr3_+_108186721
|
11.798
|
NM_001190161
NM_019976
|
Psrc1
|
proline/serine-rich coiled-coil 1
|
|
chr1_+_136859183
|
11.734
|
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative)
|
|
chr9_-_21564723
|
11.694
|
NM_026282
|
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr11_-_97049143
|
11.675
|
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chr2_-_127657532
|
11.662
|
NM_001113179
NM_009772
|
Bub1
|
budding uninhibited by benzimidazoles 1 homolog (S. cerevisiae)
|
|
chr17_+_88071837
|
11.346
|
NM_008628
|
Msh2
|
mutS homolog 2 (E. coli)
|
|
chr3_+_79433000
|
11.292
|
NM_029482
|
4930579G24Rik
|
RIKEN cDNA 4930579G24 gene
|
|
chr6_+_116214215
|
11.268
|
NM_001081317
|
Anubl1
|
AN1, ubiquitin-like, homolog (Xenopus laevis)
|
|
chr2_-_129122892
|
11.257
|
NM_181589
|
Ckap2l
|
cytoskeleton associated protein 2-like
|
|
chr3_+_134875526
|
11.197
|
NM_173762
|
Cenpe
|
centromere protein E
|
|
chr4_+_114672849
|
11.069
|
|
Stil
|
Scl/Tal1 interrupting locus
|
|
chr4_+_132324212
|
11.040
|
NM_011284
|
Rpa2
|
replication protein A2
|
|
chr2_+_118639738
|
10.867
|
NM_026412
|
D2Ertd750e
|
DNA segment, Chr 2, ERATO Doi 750, expressed
|
|
chr17_+_56442759
|
10.861
|
NM_001111079
NM_010931
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1
|
|
chr4_+_114672722
|
10.857
|
NM_009185
|
Stil
|
Scl/Tal1 interrupting locus
|
|
chr11_-_100956699
|
10.813
|
NM_008949
|
Psmc3ip
|
proteasome (prosome, macropain) 26S subunit, ATPase 3, interacting protein
|
|
chr14_-_66452720
|
10.748
|
NM_028039
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr15_-_98507886
|
10.640
|
NM_172612
|
Rnd1
|
Rho family GTPase 1
|
|
chr6_+_124780193
|
10.396
|
NM_013538
|
Cdca3
|
cell division cycle associated 3
|
|
chr6_-_126889539
|
10.381
|
NM_009013
|
Rad51ap1
|
RAD51 associated protein 1
|
|
chr10_-_87609629
|
10.258
|
NM_029249
|
4930547N16Rik
|
RIKEN cDNA 4930547N16 gene
|
|
chr10_+_87609768
|
10.230
|
NM_028334
|
Nup37
|
nucleoporin 37
|
|
chr2_+_152673699
|
10.218
|
NM_001141975
NM_001141976
NM_001141977
NM_001141978
NM_028109
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr11_-_97049185
|
10.124
|
NM_008379
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chr4_+_98590457
|
10.107
|
NM_146144
|
Usp1
|
ubiquitin specific peptidase 1
|
|
chr10_+_87609736
|
10.090
|
NM_027191
|
Nup37
|
nucleoporin 37
|
|
chrX_-_90877453
|
10.046
|
NM_008892
|
Pola1
|
polymerase (DNA directed), alpha 1
|
|
chr13_-_21879086
|
10.024
|
NM_178184
|
Hist1h2an
|
histone cluster 1, H2an
|
|
chr14_-_99498905
|
9.989
|
NM_028315
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr11_-_106860803
|
9.965
|
NM_010655
|
Kpna2
|
karyopherin (importin) alpha 2
|
|
chr11_+_98769122
|
9.910
|
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr10_+_20067606
|
9.846
|
NM_027930
|
Fam54a
|
family with sequence similarity 54, member A
|
|
chr7_-_87679873
|
9.751
|
NM_007550
NM_001042527
|
Blm
|
Bloom syndrome, RecQ helicase-like
|
|
chr17_+_27693518
|
9.723
|
NM_001166476
NM_001166477
NM_001025427
NM_001039356
NM_001166535
NM_001166536
NM_001166539
NM_001166540
NM_001166541
NM_001166542
NM_001166543
NM_001166544
NM_001166545
NM_001166546
NM_001166537
NM_016660
|
Hmga1-rs1
Hmga1
|
high mobility group AT-hook I, related sequence 1
high mobility group AT-hook 1
|
|
chr2_+_166731527
|
9.678
|
NM_023565
|
Cse1l
|
chromosome segregation 1-like (S. cerevisiae)
|
|
chr3_-_98143778
|
9.574
|
NM_016966
|
Phgdh
|
3-phosphoglycerate dehydrogenase
|
|
chr13_+_109106530
|
9.487
|
NM_178683
|
Depdc1b
|
DEP domain containing 1B
|
|
chr14_-_48038081
|
9.461
|
NM_001145949
NM_144553
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr5_+_116295259
|
9.327
|
|
Cit
|
citron
|
|
chr6_+_124780226
|
9.286
|
|
Cdca3
|
cell division cycle associated 3
|
|
chr7_+_129302950
|
9.248
|
NM_011121
|
Plk1
|
polo-like kinase 1 (Drosophila)
|
|
chr1_+_133424461
|
9.228
|
|
Fam72a
|
family with sequence similarity 72, member A
|
|
chr12_+_114394594
|
9.160
|
NM_134041
|
4930427A07Rik
|
RIKEN cDNA 4930427A07 gene
|
|
chr11_+_98769464
|
9.063
|
NM_011799
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr11_+_103828184
|
9.019
|
NM_007477
|
Arf2
|
ADP-ribosylation factor 2
|
|
chr6_-_47544925
|
8.991
|
NM_001146689
NM_007971
|
Ezh2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr14_-_87540920
|
8.647
|
NM_019670
|
Diap3
|
diaphanous homolog 3 (Drosophila)
|
|
chr11_-_40546863
|
8.563
|
NM_013552
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM)
|
|
chr14_+_68334187
|
8.459
|
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chr14_-_48037815
|
8.431
|
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr5_-_138613028
|
8.330
|
NM_008568
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae)
|
|
chr14_-_87540995
|
8.282
|
|
Diap3
|
diaphanous homolog 3 (Drosophila)
|
|
chr17_+_46633703
|
8.227
|
NM_207161
|
BC048355
|
cDNA sequence BC048355
|
|
chr18_-_34911152
|
8.191
|
NM_009860
|
Cdc25c
|
cell division cycle 25 homolog C (S. pombe)
|
|
chr7_-_59117613
|
8.147
|
NM_001115087
|
Fancf
|
Fanconi anemia, complementation group F
|
|
chr14_+_68334154
|
8.143
|
NM_001111028
NM_134073
|
Kctd9
|
potassium channel tetramerisation domain containing 9
|
|
chr1_-_191511950
|
8.117
|
NM_001081363
|
Cenpf
|
centromere protein F
|
|
chr9_-_61794540
|
8.083
|
NM_024245
|
Kif23
|
kinesin family member 23
|
|
chr11_-_61307598
|
8.012
|
NM_011841
|
Mapk7
|
mitogen-activated protein kinase 7
|
|
chr8_-_13890244
|
7.982
|
NM_025556
|
2410022L05Rik
|
RIKEN cDNA 2410022L05 gene
|
|
chr4_+_108252056
|
7.969
|
NM_011015
|
Orc1
|
origin recognition complex, subunit 1
|
|
chr10_+_7387285
|
7.907
|
NM_145706
|
Nup43
|
nucleoporin 43
|
|
chr11_-_86014640
|
7.907
|
NM_178309
|
Brip1
|
BRCA1 interacting protein C-terminal helicase 1
|
|
chr18_-_33373428
|
7.876
|
NM_133774
|
Stard4
|
StAR-related lipid transfer (START) domain containing 4
|
|
chr6_-_8209105
|
7.821
|
|
Rpa3
|
replication protein A3
|
|
chr5_+_91320265
|
7.805
|
NM_008176
|
Cxcl1
|
chemokine (C-X-C motif) ligand 1
|
|
chr13_-_23854226
|
7.803
|
NM_013550
|
Hist1h3a
|
histone cluster 1, H3a
|
|
chr14_-_48037874
|
7.782
|
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5
|
|
chr6_+_85401450
|
7.766
|
NM_007638
|
Cct7
|
chaperonin containing Tcp1, subunit 7 (eta)
|
|
chr2_+_118639930
|
7.735
|
|
D2Ertd750e
|
DNA segment, Chr 2, ERATO Doi 750, expressed
|
|
chr17_+_71554015
|
7.626
|
|
Lpin2
|
lipin 2
|
|
chr13_-_12350266
|
7.567
|
NM_001081128
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase
|
|
chr1_-_193399412
|
7.560
|
NM_029766
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr13_+_23646900
|
7.476
|
NM_145713
|
Hist1h1d
|
histone cluster 1, H1d
|
|
chr15_+_98905403
|
7.398
|
NM_001162506
NM_030159
|
Troap
|
trophinin associated protein
|
|
chr19_-_8893759
|
7.388
|
NM_027412
|
Ttc9c
|
tetratricopeptide repeat domain 9C
|
|
chr4_+_98590565
|
7.357
|
|
Usp1
|
ubiquitin specific peptidase 1
|
|
chr4_+_130037378
|
7.330
|
NM_025645
|
Snrnp40
|
small nuclear ribonucleoprotein 40 (U5)
|
|
chr6_-_8209135
|
7.294
|
NM_026632
|
Rpa3
|
replication protein A3
|
|
chr15_+_85690375
|
7.212
|
NM_013882
|
Gtse1
|
G two S phase expressed protein 1
|
|
chr6_+_128312987
|
7.185
|
NM_008021
|
Foxm1
|
forkhead box M1
|
|
chr6_+_145564482
|
7.184
|
NM_009449
|
Tuba3b
|
tubulin, alpha 3B
|
|
chr11_-_61307721
|
7.087
|
|
Mapk7
|
mitogen-activated protein kinase 7
|
|
chr6_-_125236045
|
7.073
|
NM_009446
|
Tuba3a
|
tubulin, alpha 3A
|
|
chr6_+_128313041
|
7.032
|
|
Foxm1
|
forkhead box M1
|
|
chr4_-_91042997
|
6.913
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chrX_-_137491205
|
6.881
|
NM_001164177
NM_016883
|
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10
|
|
chr13_+_49777464
|
6.849
|
NM_172015
|
Iars
|
isoleucine-tRNA synthetase
|
|
chr13_-_21872421
|
6.828
|
NM_020034
|
Hist1h1b
|
histone cluster 1, H1b
|
|
chr4_-_132066370
|
6.819
|
NM_144907
|
Sesn2
|
sestrin 2
|
|
chr13_+_23666249
|
6.729
|
NM_178188
|
Hist1h2ad
Hist1h2ai
|
histone cluster 1, H2ad
histone cluster 1, H2ai
|
|
chr17_-_71196116
|
6.687
|
NM_001164077
|
Tgif1
|
TGFB-induced factor homeobox 1
|
|
chr2_+_72314235
|
6.646
|
NM_025866
|
Cdca7
|
cell division cycle associated 7
|
|
chr7_-_27963760
|
6.616
|
NM_029391
|
Rab4b
|
RAB4B, member RAS oncogene family
|
|
chr17_+_71553920
|
6.575
|
NM_001164885
|
Lpin2
|
lipin 2
|
|
chr6_-_145024487
|
6.568
|
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr11_+_105043195
|
6.565
|
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis)
|
|
chr16_-_48994193
|
6.552
|
|
Dzip3
|
DAZ interacting protein 3, zinc finger
|
|
chr3_-_75360749
|
6.533
|
|
Pdcd10
|
programmed cell death 10
|
|
chr1_+_133424544
|
6.516
|
NM_175382
|
Fam72a
|
family with sequence similarity 72, member A
|
|
chr4_+_11486066
|
6.447
|
NM_001039556
|
Rad54b
|
RAD54 homolog B (S. cerevisiae)
|
|
chr15_+_81846798
|
6.444
|
NM_010247
|
Xrcc6
|
X-ray repair complementing defective repair in Chinese hamster cells 6
|
|
chr4_-_129318200
|
6.422
|
|
Txlna
|
taxilin alpha
|
|
chr10_-_88906629
|
6.407
|
NM_001033331
NM_001079876
|
Gas2l3
|
growth arrest-specific 2 like 3
|
|
chr15_+_85690136
|
6.403
|
NM_001168672
|
Gtse1
|
G two S phase expressed protein 1
|
|
chr6_-_85401198
|
6.386
|
NM_001163427
NM_028505
|
1700040I03Rik
|
RIKEN cDNA 1700040I03 gene
|
|
chr10_+_75495391
|
6.369
|
|
Zfp280b
|
zinc finger protein 280B
|
|
chr2_+_119177025
|
6.369
|
|
Chac1
|
ChaC, cation transport regulator-like 1 (E. coli)
|
|
chr1_+_141351340
|
6.302
|
NM_009791
|
Aspm
|
asp (abnormal spindle)-like, microcephaly associated (Drosophila)
|
|
chrX_+_132804791
|
6.289
|
NM_001110233
NM_001110234
NM_009750
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr12_+_101122706
|
6.269
|
NM_028354
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1
|
|
chr5_+_114580397
|
6.242
|
NM_001040691
|
Ung
|
uracil DNA glycosylase
|
|
chr3_-_100773571
|
6.138
|
NM_001013026
|
Ttf2
|
transcription termination factor, RNA polymerase II
|
|
chr19_-_8893358
|
6.137
|
|
Ttc9c
|
tetratricopeptide repeat domain 9C
|
|
chr13_-_34222209
|
6.136
|
NM_023716
|
Tubb2b
|
tubulin, beta 2B
|
|
chr6_-_145024451
|
6.083
|
|
Bcat1
|
branched chain aminotransferase 1, cytosolic
|
|
chr18_+_76401531
|
6.020
|
NM_010754
|
Smad2
|
MAD homolog 2 (Drosophila)
|
|
chrX_+_132804834
|
5.984
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr7_-_56136405
|
5.925
|
NM_001013368
|
E2f8
|
E2F transcription factor 8
|
|
chr6_-_13789847
|
5.914
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr5_-_4104696
|
5.884
|
NM_020010
|
Cyp51
|
cytochrome P450, family 51
|
|
chr2_+_130100145
|
5.884
|
NM_024193
|
Nop56
|
NOP56 ribonucleoprotein homolog (yeast)
|
|
chr12_-_87165448
|
5.882
|
NM_021446
|
0610007P14Rik
|
RIKEN cDNA 0610007P14 gene
|
|
chr11_+_81859212
|
5.865
|
NM_013654
|
Ccl7
|
chemokine (C-C motif) ligand 7
|
|
chrX_+_132804865
|
5.857
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr16_-_18289209
|
5.841
|
NM_033324
|
Dgcr8
|
DiGeorge syndrome critical region gene 8
|
|
chr2_+_53050461
|
5.816
|
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6
|
|
chr2_-_154395469
|
5.796
|
NM_007891
|
E2f1
|
E2F transcription factor 1
|
|
chr8_+_77633426
|
5.785
|
NM_008566
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae)
|