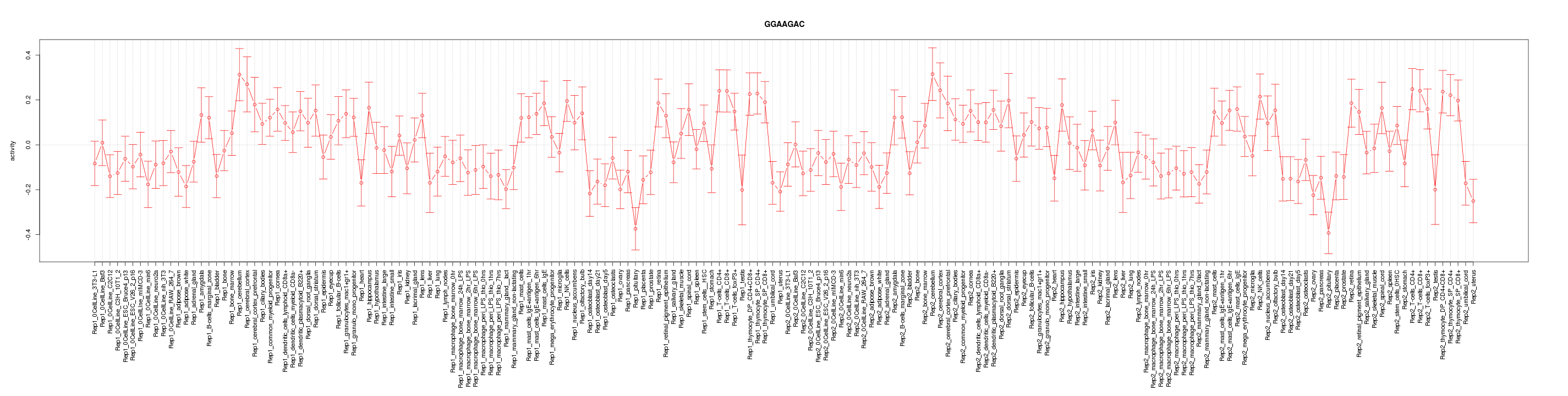
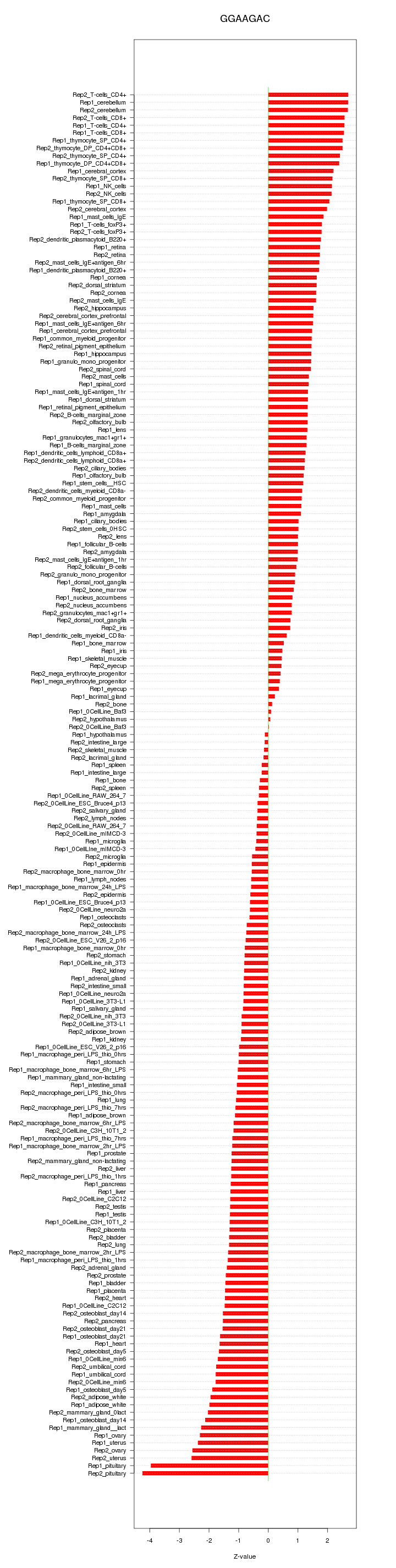
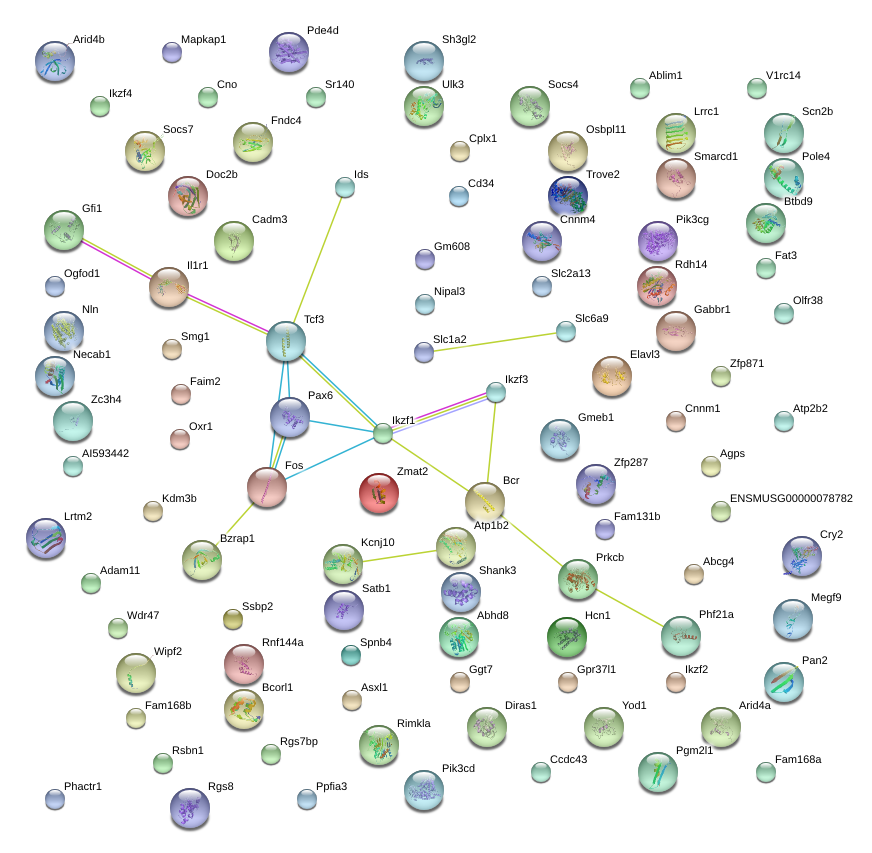
|
chr7_+_129432585
|
34.117
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr17_-_51965912
|
24.119
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51972514
|
22.796
|
NM_001163632
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51951378
|
21.069
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51971946
|
19.671
|
NM_009122
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr11_-_118770873
|
17.238
|
NM_001024931
NM_001039167
NM_001039168
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3
|
|
chr19_-_57271875
|
15.869
|
NM_001103177
|
Ablim1
|
actin-binding LIM protein 1
|
|
chr13_-_105844882
|
15.386
|
NM_029879
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr19_-_57290515
|
15.018
|
NM_178688
|
Ablim1
|
actin-binding LIM protein 1
|
|
chr13_+_91600662
|
14.499
|
NM_024186
NM_024272
|
Ssbp2
|
single-stranded DNA binding protein 2
|
|
chr1_-_175297778
|
14.140
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chr6_-_113991945
|
13.932
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_-_113841356
|
13.609
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_-_119279221
|
13.339
|
NM_001172207
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr4_-_15076093
|
13.312
|
NM_178617
|
Necab1
|
N-terminal EF-hand calcium binding protein 1
|
|
chr9_-_77392505
|
13.284
|
NM_001146048
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr9_-_77391865
|
12.454
|
NM_172528
|
Lrrc1
|
leucine rich repeat containing 1
|
|
chr13_+_118391126
|
12.351
|
NM_010408
|
Hcn1
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 1
|
|
chr4_-_70195952
|
11.745
|
NM_172694
|
Megf9
|
multiple EGF-like-domains 9
|
|
chr9_-_21856445
|
11.629
|
NM_010487
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr6_-_82602836
|
11.137
|
NM_025882
|
Pole4
|
polymerase (DNA-directed), epsilon 4 (p12 subunit)
|
|
chr13_+_109444370
|
10.918
|
NM_011056
|
Pde4d
|
phosphodiesterase 4D, cAMP specific
|
|
chr6_-_119280616
|
10.718
|
NM_172492
|
Lrtm2
|
leucine-rich repeats and transmembrane domains 2
|
|
chr11_-_75609557
|
10.316
|
NM_007873
|
Doc2b
|
double C2, beta
|
|
chr4_-_119165183
|
9.927
|
NM_177572
|
Rimkla
|
ribosomal modification protein rimK-like family member A
|
|
chr7_-_52622373
|
9.820
|
NM_029741
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr12_+_72116953
|
9.120
|
NM_001081195
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr5_-_31597792
|
9.046
|
NM_022424
|
Fndc4
|
fibronectin type III domain containing 4
|
|
chr5_-_108978950
|
9.010
|
NM_007756
|
Cplx1
|
complexin 1
|
|
chr3_+_103718042
|
8.993
|
NM_172684
|
Rsbn1
|
rosbin, round spermatid basic protein 1
|
|
chr1_+_155500166
|
8.839
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chr11_+_98724908
|
8.838
|
NM_197940
|
Wipf2
|
WAS/WASL interacting protein family, member 2
|
|
chr16_+_44173356
|
8.703
|
NM_001029889
|
Gm608
|
predicted gene 608
|
|
chr11_+_102622745
|
8.410
|
NM_001110778
NM_009613
|
Adam11
|
a disintegrin and metallopeptidase domain 11
|
|
chr2_+_75670230
|
8.185
|
NM_172666
|
Agps
|
alkylglycerone phosphate synthase
|
|
chr1_+_36528441
|
8.158
|
NM_033570
|
Cnnm4
|
cyclin M4
|
|
chr15_+_99532698
|
7.836
|
NM_031842
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr7_+_107855215
|
7.799
|
NM_178764
|
Fam168a
|
family with sequence similarity 168, member A
|
|
chr19_-_57193391
|
7.709
|
NM_001103178
|
Ablim1
|
actin-binding LIM protein 1
|
|
chr4_-_131817388
|
7.419
|
NM_001122992
NM_020273
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1
|
|
chr2_+_105516601
|
7.186
|
NM_013627
|
Pax6
|
paired box gene 6
|
|
chr9_+_44925922
|
6.900
|
NM_001014761
|
Scn2b
|
sodium channel, voltage-gated, type II, beta
|
|
chr18_+_36953553
|
6.879
|
NM_025594
|
Zmat2
|
zinc finger, matrin type 2
|
|
chr1_-_34899878
|
6.716
|
NM_001160235
NM_001160236
NM_174997
|
Fam168b
|
family with sequence similarity 168, member B
|
|
chr15_-_99358387
|
6.645
|
NM_001038658
NM_028224
|
Faim2
|
Fas apoptotic inhibitory molecule 2
|
|
chr11_-_62542594
|
6.594
|
NM_133208
|
Zfp287
|
zinc finger protein 287
|
|
chr6_-_42274556
|
6.482
|
NM_001113327
NM_029528
|
Fam131b
|
family with sequence similarity 131, member B
|
|
chr15_+_41662472
|
6.429
|
NM_001130164
|
Oxr1
3110047M12Rik
|
oxidation resistance 1
RIKEN cDNA 3110047M12 gene
|
|
chr7_+_107376116
|
6.403
|
NM_027629
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr9_-_95412264
|
6.142
|
NM_001114977
NM_026476
|
2610101N10Rik
|
RIKEN cDNA 2610101N10 gene
|
|
chr18_+_34936642
|
6.034
|
NM_001081256
|
Kdm3b
|
KDM3B lysine (K)-specific demethylase 3B
|
|
chr2_+_153171730
|
5.902
|
NM_001039939
|
Asxl1
|
additional sex combs like 1 (Drosophila)
|
|
chr15_+_41278949
|
5.840
|
NM_001130166
|
Oxr1
|
oxidation resistance 1
|
|
chr7_-_125386862
|
5.819
|
NM_001031814
|
Smg1
|
SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans)
|
|
chrX_-_67618235
|
5.795
|
NM_010498
|
Ids
|
iduronate 2-sulfatase
|
|
chr16_+_33185137
|
5.576
|
NM_176840
|
Osbpl11
|
oxysterol binding protein-like 11
|
|
chr11_+_87574041
|
5.452
|
NM_172449
|
Bzrap1
|
benzodiazepine receptor associated protein 1
|
|
chr10_+_127740390
|
5.371
|
NM_133992
|
Pan2
|
PAN2 polyA specific ribonuclease subunit homolog (S. cerevisiae)
|
|
chr16_+_5884885
|
5.350
|
NM_021477
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr12_-_27100112
|
5.349
|
NM_001081977
NM_080563
|
Rnf144a
|
ring finger protein 144A
|
|
chr7_+_16986478
|
5.196
|
NM_198631
|
Zc3h4
|
zinc finger CCCH-type containing 4
|
|
chr9_+_57437238
|
5.175
|
NM_027895
|
Ulk3
|
unc-51-like kinase 3 (C. elegans)
|
|
chr4_-_149075697
|
5.129
|
NM_001029837
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr4_-_149050154
|
5.115
|
NM_001164051
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr2_-_92274110
|
4.945
|
NM_001113333
NM_009963
|
Cry2
|
cryptochrome 2 (photolyase-like)
|
|
chr12_+_86814822
|
4.811
|
NM_010234
|
Fos
|
FBJ osteosarcoma oncogene
|
|
chr9_-_16182674
|
4.797
|
NM_001080814
|
Fat3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr15_+_41620484
|
4.737
|
NM_001130165
|
Oxr1
|
oxidation resistance 1
|
|
chr16_+_7069927
|
4.729
|
NM_183188
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr15_+_41620926
|
4.712
|
NM_001130163
NM_130885
|
Oxr1
|
oxidation resistance 1
|
|
chrX_+_45694534
|
4.609
|
NM_178782
|
Bcorl1
|
BCL6 co-repressor-like 1
|
|
chr12_-_32893074
|
4.485
|
NM_001146201
NM_020272
NM_001146200
|
Pik3cg
|
phosphoinositide-3-kinase, catalytic, gamma polypeptide
|
|
chr4_+_117507819
|
4.357
|
NM_008135
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9
|
|
chr2_+_102498839
|
4.355
|
NM_001077514
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr5_-_37139883
|
4.283
|
NM_133724
|
Cno
|
cappuccino
|
|
chr9_-_44096170
|
4.237
|
NM_138955
|
Abcg4
|
ATP-binding cassette, sub-family G (WHITE), member 4
|
|
chr2_+_34287544
|
4.132
|
NM_177345
|
Mapkap1
|
mitogen-activated protein kinase associated protein 1
|
|
chr19_+_43514902
|
4.105
|
NM_031396
|
Cnnm1
|
cyclin M1
|
|
chr11_+_97223851
|
4.093
|
NM_138657
|
Socs7
|
suppressor of cytokine signaling 7
|
|
chr1_-_137064124
|
4.083
|
NM_134438
|
Gpr37l1
|
G protein-coupled receptor 37-like 1
|
|
chr10_-_80488111
|
4.024
|
NM_145217
|
Diras1
|
DIRAS family, GTP-binding RAS-like 1
|
|
chr1_+_174271338
|
4.018
|
NM_001039484
|
Kcnj10
|
potassium inwardly-rectifying channel, subfamily J, member 10
|
|
chr5_-_108153337
|
3.906
|
NM_010278
|
Gfi1
|
growth factor independent 1
|
|
chr13_+_42775991
|
3.879
|
NM_198419
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr11_-_102559011
|
3.864
|
NM_025918
|
Ccdc43
|
coiled-coil domain containing 43
|
|
chr15_+_89330287
|
3.786
|
NM_021423
|
Shank3
|
SH3/ankyrin domain gene 3
|
|
chr17_-_32925218
|
3.772
|
NM_172458
|
Zfp871
|
zinc finger protein 871
|
|
chr11_-_69419419
|
3.753
|
NM_013415
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr3_+_108394195
|
3.738
|
NM_181400
|
Wdr47
|
WD repeat domain 47
|
|
chr13_+_43218207
|
3.696
|
NM_001005748
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr8_-_73987384
|
3.663
|
NM_022419
|
Abhd8
|
abhydrolase domain containing 8
|
|
chr11_+_11586215
|
3.661
|
NM_001025597
NM_009578
|
Ikzf1
|
IKAROS family zinc finger 1
|
|
chr4_+_84851330
|
3.641
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr9_-_52487410
|
3.567
|
NM_178906
|
AI593442
|
expressed sequence AI593442
|
|
chr17_-_30713158
|
3.534
|
NM_027060
NM_172618
|
Btbd9
|
BTB (POZ) domain containing 9
|
|
chr13_+_42804911
|
3.505
|
NM_001005740
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr8_+_96561097
|
3.495
|
NM_001093757
NM_177767
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1
|
|
chr7_-_28231535
|
3.459
|
NM_032610
|
Spnb4
|
spectrin beta 4
|
|
chr1_+_132613903
|
3.393
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr4_-_135050358
|
3.365
|
NM_028995
|
Nipal3
|
NIPA-like domain containing 3
|
|
chr17_+_37182910
|
3.362
|
NM_019439
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1
|
|
chr2_+_92025639
|
3.279
|
NM_001109691
|
Phf21a
|
PHD finger protein 21A
|
|
chr2_-_155340538
|
3.257
|
NM_144786
|
Ggt7
|
gamma-glutamyltransferase 7
|
|
chr10_+_74523633
|
3.254
|
NM_001081412
|
Bcr
|
breakpoint cluster region
|
|
chr8_+_107064705
|
3.251
|
NM_053070
|
Car7
|
carbonic anhydrase 7
|
|
chr18_-_60769648
|
3.219
|
NM_001109975
|
Synpo
|
synaptopodin
|
|
chr18_-_15562126
|
3.191
|
NM_009700
|
Aqp4
|
aquaporin 4
|
|
chr12_-_82881995
|
3.179
|
NM_001174107
NM_177395
|
Map3k9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr2_+_158201373
|
3.178
|
NM_175692
|
Snhg11
|
small nucleolar RNA host gene 11
|
|
chr6_+_114081227
|
3.170
|
NM_172890
|
Slc6a11
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 11
|
|
chr10_+_34017221
|
3.146
|
NM_030203
|
Tspyl4
|
TSPY-like 4
|
|
chr8_+_13869640
|
3.108
|
NM_181854
|
Zfp828
|
zinc finger protein 828
|
|
chr2_+_154959216
|
3.100
|
NM_008395
|
Itch
|
itchy, E3 ubiquitin protein ligase
|
|
chr6_-_126595818
|
3.098
|
NM_010595
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1
|
|
chr16_+_3884618
|
3.068
|
NM_029090
|
Nat15
|
N-acetyltransferase 15 (GCN5-related, putative)
|
|
chr7_+_94732677
|
3.038
|
NM_001081414
NM_001143834
|
Grm5
|
glutamate receptor, metabotropic 5
|
|
chr5_+_136293975
|
3.032
|
NM_021403
|
Srrm3
|
serine/arginine repetitive matrix 3
|
|
chr6_-_57485419
|
3.030
|
NM_175523
|
Ppm1k
|
protein phosphatase 1K (PP2C domain containing)
|
|
chr10_+_90038471
|
3.017
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr17_+_74154180
|
3.003
|
NM_020578
|
Ehd3
|
EH-domain containing 3
|
|
chr17_-_66426361
|
2.916
|
NM_001025572
|
Ankrd12
|
ankyrin repeat domain 12
|
|
chr5_+_107926763
|
2.876
|
NM_001007574
NM_001168557
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene
|
|
chr9_+_64658817
|
2.873
|
NM_001162917
|
Dennd4a
|
DENN/MADD domain containing 4A
|
|
chr1_+_40522396
|
2.867
|
NM_001161842
NM_001161843
|
Il18r1
|
interleukin 18 receptor 1
|
|
chr7_-_116925058
|
2.833
|
NM_020610
|
Nrip3
|
nuclear receptor interacting protein 3
|
|
chr1_+_60466462
|
2.812
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr9_-_40965548
|
2.800
|
NM_176860
|
Ubash3b
|
ubiquitin associated and SH3 domain containing, B
|
|
chr6_+_118016385
|
2.728
|
NM_027526
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr14_-_9142289
|
2.703
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr18_+_61123204
|
2.666
|
NM_009792
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr18_+_65590615
|
2.624
|
NM_172833
|
Malt1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1
|
|
chr19_+_11986887
|
2.622
|
NM_172635
|
Patl1
|
protein associated with topoisomerase II homolog 1 (yeast)
|
|
chr1_-_133141708
|
2.591
|
NM_018750
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr8_-_17535279
|
2.587
|
NM_053171
|
Csmd1
|
CUB and Sushi multiple domains 1
|
|
chr18_-_74224541
|
2.584
|
NM_172632
|
Mapk4
|
mitogen-activated protein kinase 4
|
|
chr4_+_134676559
|
2.583
|
NM_019732
|
Runx3
|
runt related transcription factor 3
|
|
chr2_+_92061718
|
2.570
|
NM_001109690
|
Phf21a
|
PHD finger protein 21A
|
|
chr7_+_106683983
|
2.558
|
NM_177231
NM_178220
|
Arrb1
|
arrestin, beta 1
|
|
chr14_-_55735114
|
2.553
|
NM_177049
|
Jph4
|
junctophilin 4
|
|
chr4_-_149072755
|
2.544
|
NM_001164049
NM_001164050
NM_008840
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr11_-_70501586
|
2.480
|
NM_001190376
NM_001190378
NM_001190379
NM_178116
|
Camta2
|
calmodulin binding transcription activator 2
|
|
chr11_+_69579376
|
2.465
|
NM_029348
|
Zbtb4
|
zinc finger and BTB domain containing 4
|
|
chr2_+_102546576
|
2.465
|
NM_001077515
NM_011393
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2
|
|
chr11_-_86358252
|
2.462
|
NM_001114334
NM_028259
|
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1
|
|
chr10_+_89336253
|
2.444
|
NM_001128086
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr4_-_107973683
|
2.400
|
NM_001033634
|
Zyg11b
|
zyg-ll homolog B (C. elegans)
|
|
chr9_-_62828677
|
2.351
|
NM_019663
|
Pias1
|
protein inhibitor of activated STAT 1
|
|
chr3_+_103083215
|
2.325
|
NM_001079830
NM_053170
|
Trim33
|
tripartite motif-containing 33
|
|
chr14_-_27783615
|
2.312
|
NM_145221
|
Appl1
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1
|
|
chr6_-_112897259
|
2.290
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr14_+_31638767
|
2.282
|
NM_028839
|
Tmem110
|
transmembrane protein 110
|
|
chr9_-_83699706
|
2.280
|
NM_001145974
NM_148941
|
Elovl4
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4
|
|
chr9_+_43851466
|
2.262
|
NM_009382
|
Thy1
|
thymus cell antigen 1, theta
|
|
chr12_-_68323528
|
2.256
|
NM_001193266
NM_207010
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2
|
|
chr2_+_164793487
|
2.251
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr7_+_134072351
|
2.237
|
NM_172747
|
Kctd13
|
potassium channel tetramerisation domain containing 13
|
|
chr1_-_172236462
|
2.229
|
NM_001198955
|
Gm7694
|
predicted gene 7694
|
|
chr10_+_101837068
|
2.228
|
NM_026243
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative)
|
|
chr15_+_100766701
|
2.225
|
NM_011323
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr2_-_132211105
|
2.222
|
NM_144944
|
Prokr2
|
prokineticin receptor 2
|
|
chr1_-_162281701
|
2.208
|
NM_001038621
|
Rabgap1l
|
RAB GTPase activating protein 1-like
|
|
chr9_-_105034128
|
2.187
|
NM_029385
|
Nudt16
|
nudix (nucleoside diphosphate linked moiety X)-type motif 16
|
|
chr4_+_135451278
|
2.179
|
NM_009924
|
Cnr2
|
cannabinoid receptor 2 (macrophage)
|
|
chr9_-_75446888
|
2.177
|
NM_001038710
|
Tmod2
|
tropomodulin 2
|
|
chr3_+_156224757
|
2.169
|
NM_001039094
NM_177274
|
Negr1
|
neuronal growth regulator 1
|
|
chr9_-_75459125
|
2.162
|
NM_016711
|
Tmod2
|
tropomodulin 2
|
|
chr12_-_119539909
|
2.152
|
NM_001166385
NM_009239
|
Sp4
|
trans-acting transcription factor 4
|
|
chrX_+_157205944
|
2.146
|
NM_011302
|
Rs1
|
retinoschisis (X-linked, juvenile) 1 (human)
|
|
chr11_+_110927475
|
2.120
|
NM_008425
|
Kcnj2
|
potassium inwardly-rectifying channel, subfamily J, member 2
|
|
chr9_+_107838164
|
2.108
|
NM_145621
|
Camkv
|
CaM kinase-like vesicle-associated
|
|
chr6_+_134590957
|
2.056
|
NM_026371
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human)
|
|
chr1_-_74931895
|
2.045
|
NM_153111
|
Fev
|
FEV (ETS oncogene family)
|
|
chr1_+_162836541
|
2.045
|
NM_001024952
|
Rc3h1
|
RING CCCH (C3H) domains 1
|
|
chr10_+_85061155
|
2.043
|
NM_001017525
|
Btbd11
|
BTB (POZ) domain containing 11
|
|
chr17_+_34375847
|
2.038
|
NM_010389
|
H2-Ob
|
histocompatibility 2, O region beta locus
|
|
chrX_-_34529439
|
2.038
|
NM_001177323
|
Sept6
|
septin 6
|
|
chr1_-_58642994
|
2.019
|
NM_172513
|
Fam126b
|
family with sequence similarity 126, member B
|
|
chr4_+_151712739
|
2.016
|
NM_001081376
NM_029216
|
Chd5
|
chromodomain helicase DNA binding protein 5
|
|
chr14_+_37868015
|
2.016
|
NM_146245
|
Lrit1
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1
|
|
chr11_-_69648350
|
2.009
|
NM_198862
|
Nlgn2
|
neuroligin 2
|
|
chr6_+_86354212
|
2.006
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr9_-_96531911
|
2.005
|
NM_053268
|
Rasa2
|
RAS p21 protein activator 2
|
|
chr16_-_45654180
|
2.001
|
NM_145389
|
BC016579
|
cDNA sequence, BC016579
|
|
chr17_-_37160241
|
1.993
|
NM_010814
|
Mog
|
myelin oligodendrocyte glycoprotein
|
|
chr6_+_8459595
|
1.982
|
NM_133236
|
Glcci1
|
glucocorticoid induced transcript 1
|
|
chr7_+_51846254
|
1.962
|
NM_008422
|
Kcnc3
|
potassium voltage gated channel, Shaw-related subfamily, member 3
|
|
chr7_-_25790888
|
1.961
|
NM_144921
|
Atp1a3
|
ATPase, Na+/K+ transporting, alpha 3 polypeptide
|
|
chr1_-_17087821
|
1.956
|
NM_020604
|
Jph1
|
junctophilin 1
|
|
chr2_-_180773777
|
1.954
|
NM_015730
|
Chrna4
|
cholinergic receptor, nicotinic, alpha polypeptide 4
|
|
chr2_-_51790163
|
1.922
|
NM_001141982
|
Rbm43
|
RNA binding motif protein 43
|
|
chr6_+_8159188
|
1.891
|
NM_145374
|
Mios
|
missing oocyte, meiosis regulator, homolog (Drosophila)
|
|
chr1_+_133807034
|
1.890
|
NM_001145804
NM_175294
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1
|
|
chr15_+_100701068
|
1.883
|
NM_001077499
|
Scn8a
|
sodium channel, voltage-gated, type VIII, alpha
|
|
chr16_+_32644728
|
1.872
|
NM_001110147
NM_016788
|
Tnk2
|
tyrosine kinase, non-receptor, 2
|
|
chr6_-_85283412
|
1.861
|
NM_178639
|
Sfxn5
|
sideroflexin 5
|
|
chr2_-_51790406
|
1.860
|
NM_001141981
NM_030243
|
Rbm43
|
RNA binding motif protein 43
|
|
chr6_+_49269351
|
1.859
|
NM_175098
|
Ccdc126
|
coiled-coil domain containing 126
|
|
chr11_+_92960603
|
1.853
|
NM_028296
|
Car10
|
carbonic anhydrase 10
|
|
chr7_+_146132386
|
1.836
|
NM_028708
|
Jakmip3
|
janus kinase and microtubule interacting protein 3
|
|
chr8_-_70342105
|
1.811
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|