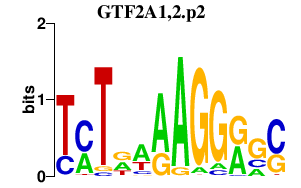
|
chrX_-_71470766
|
21.839
|
|
Flna
|
filamin, alpha
|
|
chr7_-_26262552
|
18.439
|
NM_001039185
NM_001039186
NM_001039187
NM_011926
|
Ceacam1
|
carcinoembryonic antigen-related cell adhesion molecule 1
|
|
chr11_+_94797565
|
17.527
|
NM_007742
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr1_+_90062617
|
16.682
|
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr11_-_83391984
|
16.074
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr19_+_9091248
|
14.911
|
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin)
|
|
chr11_-_83392109
|
14.535
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr1_+_132723258
|
13.630
|
NM_011082
|
Pigr
|
polymeric immunoglobulin receptor
|
|
chr1_+_132723277
|
13.215
|
|
Pigr
|
polymeric immunoglobulin receptor
|
|
chr3_+_92019756
|
13.022
|
NM_001164787
NM_011468
|
Sprr2a2
Sprr2a1
|
small proline-rich protein 2A2
small proline-rich protein 2A1
|
|
chr3_+_88128944
|
12.553
|
NM_020003
|
0610031J06Rik
|
RIKEN cDNA 0610031J06 gene
|
|
chr3_-_88307198
|
12.398
|
|
Lmna
|
lamin A
|
|
chr2_+_117937623
|
11.910
|
NM_011580
|
Thbs1
|
thrombospondin 1
|
|
chr6_-_82724366
|
11.767
|
NM_013820
|
Hk2
|
hexokinase 2
|
|
chr3_-_88307253
|
11.505
|
NM_001002011
NM_001111102
|
Lmna
|
lamin A
|
|
chr11_+_75469516
|
11.383
|
NM_001080775
|
Myo1c
|
myosin IC
|
|
chr1_-_134036041
|
11.231
|
NM_008795
|
Cdk18
|
cyclin-dependent kinase 18
|
|
chr2_+_35159780
|
10.881
|
|
Gsn
|
gelsolin
|
|
chr11_-_3429909
|
10.723
|
|
Smtn
|
smoothelin
|
|
chr7_-_52715059
|
10.720
|
NM_010240
|
Gm15590
Ftl1
Ftl2
|
ferritin light chain 1 pseudogene
ferritin light chain 1
ferritin light chain 2
|
|
chr11_+_120353150
|
10.719
|
NM_013770
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10
|
|
chr3_-_88307024
|
10.717
|
|
Lmna
|
lamin A
|
|
chr5_-_137548123
|
10.031
|
NM_008871
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr7_-_31861665
|
9.951
|
NM_008557
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3
|
|
chr17_-_31266539
|
9.626
|
NM_011575
|
Tff3
|
trefoil factor 3, intestinal
|
|
chr19_+_9090816
|
9.088
|
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin)
|
|
chr7_-_19857121
|
8.819
|
|
Vasp
|
vasodilator-stimulated phosphoprotein
|
|
chr3_-_96044172
|
8.643
|
NM_013549
NM_178212
|
Hist2h2aa1
Hist2h2aa2
|
histone cluster 2, H2aa1
histone cluster 2, H2aa2
|
|
chr11_+_120353171
|
8.520
|
|
Slc25a10
|
solute carrier family 25 (mitochondrial carrier, dicarboxylate transporter), member 10
|
|
chr3_-_69663781
|
8.507
|
NM_001164210
NM_133675
|
1110032A04Rik
|
RIKEN cDNA 1110032A04 gene
|
|
chr10_+_51200463
|
8.251
|
NM_008147
|
Gp49a
|
glycoprotein 49 A
|
|
chr5_+_31215857
|
8.226
|
|
Emilin1
|
elastin microfibril interfacer 1
|
|
chr4_+_135519640
|
8.191
|
|
Gale
|
galactose-4-epimerase, UDP
|
|
chr14_+_70945119
|
8.187
|
|
Reep4
|
receptor accessory protein 4
|
|
chr19_-_4269096
|
8.170
|
|
Ssh3
|
slingshot homolog 3 (Drosophila)
|
|
chr11_-_114795651
|
7.960
|
NM_199221
|
Cd300lb
|
CD300 antigen like family member B
|
|
chr4_+_118102305
|
7.951
|
NM_001039176
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr7_-_19857195
|
7.689
|
NM_009499
|
Vasp
|
vasodilator-stimulated phosphoprotein
|
|
chr5_+_32438843
|
7.651
|
NM_008037
|
Fosl2
|
fos-like antigen 2
|
|
chr6_-_83486194
|
7.642
|
NM_009610
|
Actg2
|
actin, gamma 2, smooth muscle, enteric
|
|
chr7_+_25378887
|
7.641
|
|
Ethe1
|
ethylmalonic encephalopathy 1
|
|
chr7_-_108971734
|
7.626
|
|
|
|
|
chr7_-_53176582
|
7.615
|
NM_010129
|
Emp3
|
epithelial membrane protein 3
|
|
chr3_+_88420058
|
7.380
|
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr19_-_45824047
|
7.267
|
NM_008723
|
Npm3
|
nucleoplasmin 3
|
|
chr3_+_90416815
|
7.149
|
NM_011313
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chr2_+_14096150
|
7.147
|
NM_001081310
|
Fam23a
|
family with sequence similarity 23, member A
|
|
chr4_+_104829951
|
7.060
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr6_-_89312545
|
7.027
|
NM_008881
|
Plxna1
|
plexin A1
|
|
chr11_-_83392129
|
6.969
|
NM_011338
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr7_-_52765714
|
6.678
|
NM_001163662
NM_008749
|
Nucb1
|
nucleobindin 1
|
|
chr17_-_26005680
|
6.604
|
NM_026633
|
Fam195a
|
family with sequence similarity 195, member A
|
|
chr19_-_41068625
|
6.572
|
|
Blnk
|
B-cell linker
|
|
chr15_+_61817102
|
6.543
|
|
Myc
|
myelocytomatosis oncogene
|
|
chr12_+_81891479
|
6.537
|
NM_178682
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene
|
|
chr1_-_133176085
|
6.470
|
NM_019777
|
Ikbke
|
inhibitor of kappaB kinase epsilon
|
|
chr11_-_83406475
|
6.211
|
NM_009139
|
Ccl6
|
chemokine (C-C motif) ligand 6
|
|
chr13_-_23775827
|
6.197
|
NM_178189
|
Hist1h2ac
|
histone cluster 1, H2ac
|
|
chr4_+_104829963
|
6.125
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr6_+_52195030
|
6.033
|
|
Hoxa11as
|
HOXA11 antisense RNA (non-protein coding)
|
|
chr13_-_21925343
|
5.952
|
NM_178185
NM_001177544
|
Hist1h2ah
Hist1h2ap
Hist1h2ai
Hist1h2ao
|
histone cluster 1, H2ah
histone cluster 1, H2ap
histone cluster 1, H2ai
histone cluster 1, H2ao
|
|
chr2_+_122463622
|
5.861
|
NM_001004174
|
AA467197
|
expressed sequence AA467197
|
|
chr2_-_26789021
|
5.776
|
NM_011512
|
Surf4
|
surfeit gene 4
|
|
chr19_-_23522811
|
5.765
|
NM_174857
|
Mamdc2
|
MAM domain containing 2
|
|
chr15_+_101974978
|
5.764
|
|
Igfbp6
|
insulin-like growth factor binding protein 6
|
|
chr7_+_119571219
|
5.760
|
NM_020606
|
Parva
|
parvin, alpha
|
|
chrX_+_160811667
|
5.715
|
|
Figf
|
c-fos induced growth factor
|
|
chr3_-_145312928
|
5.706
|
NM_010516
|
Cyr61
|
cysteine rich protein 61
|
|
chr3_-_144695718
|
5.705
|
NM_017474
|
Clca3
|
chloride channel calcium activated 3
|
|
chr5_+_144165615
|
5.680
|
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr13_-_55932570
|
5.628
|
|
Pitx1
|
paired-like homeodomain transcription factor 1
|
|
chr5_+_144165482
|
5.616
|
NM_025841
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2
|
|
chr2_-_102741661
|
5.611
|
|
Cd44
|
CD44 antigen
|
|
chr11_+_88860710
|
5.564
|
NM_009546
|
Trim25
|
tripartite motif-containing 25
|
|
chr15_-_79233664
|
5.558
|
NM_172608
|
Tmem184b
|
transmembrane protein 184b
|
|
chr1_+_90034940
|
5.555
|
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A
|
|
chr17_-_26005645
|
5.528
|
|
Fam195a
|
family with sequence similarity 195, member A
|
|
chr2_+_152934184
|
5.470
|
NM_001172117
NM_010407
|
Hck
|
hemopoietic cell kinase
|
|
chr7_-_19856882
|
5.452
|
|
Vasp
|
vasodilator-stimulated phosphoprotein
|
|
chr19_+_4097894
|
5.416
|
|
Cdk2ap2
|
CDK2-associated protein 2
|
|
chr5_-_104419233
|
5.389
|
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr5_+_93238413
|
5.383
|
NM_001077595
|
Shroom3
|
shroom family member 3
|
|
chr19_+_4097344
|
5.377
|
NM_026373
|
Cdk2ap2
|
CDK2-associated protein 2
|
|
chr13_-_62959656
|
5.330
|
NM_007994
|
Fbp2
|
fructose bisphosphatase 2
|
|
chr2_+_128604653
|
5.328
|
|
Mertk
|
c-mer proto-oncogene tyrosine kinase
|
|
chr12_+_81891598
|
5.282
|
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene
|
|
chr14_+_70945429
|
5.272
|
NM_180588
|
Reep4
|
receptor accessory protein 4
|
|
chr10_-_80813039
|
5.190
|
|
Hmg20b
|
high mobility group 20 B
|
|
chr4_-_140695648
|
5.187
|
NM_001025608
NM_174996
|
D4Ertd22e
|
DNA segment, Chr 4, ERATO Doi 22, expressed
|
|
chr12_+_70473032
|
5.183
|
NM_007481
|
Arf6
|
ADP-ribosylation factor 6
|
|
chr1_+_89967340
|
5.153
|
NM_201644
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9
|
|
chr10_-_80813198
|
5.121
|
NM_001163166
|
Hmg20b
|
high mobility group 20 B
|
|
chr19_+_6276685
|
5.120
|
|
Ehd1
|
EH-domain containing 1
|
|
chr13_+_75977323
|
5.116
|
NM_053108
|
Glrx
|
glutaredoxin
|
|
chr11_+_96184439
|
5.074
|
NM_001079869
|
Hoxb3
|
homeobox B3
|
|
chr13_-_55932612
|
5.068
|
|
Pitx1
|
paired-like homeodomain transcription factor 1
|
|
chr19_-_4269161
|
5.060
|
NM_198113
|
Ssh3
|
slingshot homolog 3 (Drosophila)
|
|
chr2_-_26788845
|
5.026
|
|
Surf4
|
surfeit gene 4
|
|
chr19_-_11893431
|
5.015
|
|
Stx3
|
syntaxin 3
|
|
chr1_+_61685397
|
5.007
|
NM_001081050
|
Pard3b
|
par-3 partitioning defective 3 homolog B (C. elegans)
|
|
chr2_+_152695336
|
5.006
|
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis)
|
|
chr3_+_88420105
|
4.999
|
NM_001198911
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr5_+_31216069
|
4.982
|
|
Emilin1
|
elastin microfibril interfacer 1
|
|
chr14_+_56159204
|
4.947
|
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial)
|
|
chr1_-_146096156
|
4.924
|
NM_015811
|
Rgs1
|
regulator of G-protein signaling 1
|
|
chr4_+_104830022
|
4.883
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr2_-_92299870
|
4.883
|
NM_211358
|
Slc35c1
|
solute carrier family 35, member C1
|
|
chr17_+_35035395
|
4.830
|
NM_147151
|
Ehmt2
|
euchromatic histone lysine N-methyltransferase 2
|
|
chr4_-_118966945
|
4.807
|
NM_011732
|
Ybx1
|
Y box protein 1
|
|
chr5_+_31216131
|
4.806
|
NM_133918
|
Emilin1
|
elastin microfibril interfacer 1
|
|
chr10_-_80813067
|
4.742
|
NM_001163165
NM_010440
|
Hmg20b
|
high mobility group 20 B
|
|
chr16_+_97578333
|
4.689
|
NM_019517
|
Bace2
|
beta-site APP-cleaving enzyme 2
|
|
chr5_-_139470785
|
4.673
|
|
Pdgfa
|
platelet derived growth factor, alpha
|
|
chr1_-_73975053
|
4.656
|
|
Tns1
|
tensin 1
|
|
chr9_+_107471447
|
4.629
|
NM_010489
|
Hyal2
|
hyaluronoglucosaminidase 2
|
|
chr13_-_62959751
|
4.616
|
|
Fbp2
|
fructose bisphosphatase 2
|
|
chr10_+_43298969
|
4.582
|
NM_009846
|
Cd24a
|
CD24a antigen
|
|
chr3_+_90417072
|
4.575
|
|
S100a6
|
S100 calcium binding protein A6 (calcyclin)
|
|
chr1_+_74455956
|
4.532
|
NM_009509
|
Vil1
|
villin 1
|
|
chr1_+_89951985
|
4.513
|
NM_201641
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10
|
|
chr2_-_148269170
|
4.509
|
NM_010740
|
Cd93
|
CD93 antigen
|
|
chr15_+_78707972
|
4.472
|
|
Gga1
|
golgi associated, gamma adaptin ear containing, ARF binding protein 1
|
|
chr6_-_86743518
|
4.443
|
NM_013471
|
Anxa4
|
annexin A4
|
|
chr9_-_43047862
|
4.425
|
NM_178644
|
Oaf
|
OAF homolog (Drosophila)
|
|
chr2_+_29725034
|
4.381
|
|
Cercam
|
cerebral endothelial cell adhesion molecule
|
|
chr5_+_137463446
|
4.373
|
|
Plod3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3
|
|
chr9_+_20820403
|
4.362
|
NM_010493
|
Icam1
|
intercellular adhesion molecule 1
|
|
chr2_+_29725007
|
4.331
|
NM_207298
|
Cercam
|
cerebral endothelial cell adhesion molecule
|
|
chr11_+_70470701
|
4.311
|
NM_001136062
|
Eno3
|
enolase 3, beta muscle
|
|
chr11_+_32247758
|
4.292
|
NM_177364
|
Sh3pxd2b
|
SH3 and PX domains 2B
|
|
chr5_+_91332895
|
4.281
|
NM_009140
|
Cxcl2
|
chemokine (C-X-C motif) ligand 2
|
|
chr6_+_115372083
|
4.268
|
NM_011146
|
Pparg
|
peroxisome proliferator activated receptor gamma
|
|
chr7_+_28856254
|
4.268
|
NM_001122603
|
Fcgbp
|
Fc fragment of IgG binding protein
|
|
chr5_-_139470895
|
4.250
|
NM_008808
|
Pdgfa
|
platelet derived growth factor, alpha
|
|
chr5_-_115594042
|
4.216
|
|
Mlec
|
malectin
|
|
chr19_+_8894752
|
4.215
|
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2
|
|
chr10_+_19928244
|
4.212
|
NM_001101561
|
Gm6251
|
predicted gene 6251
|
|
chr2_-_102741483
|
4.118
|
|
Cd44
|
CD44 antigen
|
|
chr7_+_51066659
|
4.107
|
|
Klk7
|
kallikrein related-peptidase 7 (chymotryptic, stratum corneum)
|
|
chr9_+_62705890
|
4.093
|
NM_001102468
NM_138304
|
Calml4
|
calmodulin-like 4
|
|
chr10_-_128063522
|
4.092
|
NM_013765
|
Rps26
|
ribosomal protein S26
|
|
chr7_-_128217980
|
4.070
|
NM_030691
|
Igsf6
|
immunoglobulin superfamily, member 6
|
|
chr10_-_120851345
|
4.036
|
|
Rassf3
|
Ras association (RalGDS/AF-6) domain family member 3
|
|
chr5_-_145118758
|
4.026
|
|
Baiap2l1
|
BAI1-associated protein 2-like 1
|
|
chr1_-_156946739
|
4.003
|
NM_010500
|
Ier5
|
immediate early response 5
|
|
chr4_+_109088251
|
3.948
|
NM_001145948
|
Ttc39a
|
tetratricopeptide repeat domain 39A
|
|
chr2_+_103810274
|
3.939
|
NM_001142335
NM_008505
|
Lmo2
|
LIM domain only 2
|
|
chr3_-_116415169
|
3.939
|
NM_144902
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3
|
|
chr2_-_84316891
|
3.925
|
NM_001177319
NM_001177320
|
Tfpi
|
tissue factor pathway inhibitor
|
|
chr2_+_156665812
|
3.913
|
NM_173396
|
Tgif2
|
TGFB-induced factor homeobox 2
|
|
chr2_-_59963586
|
3.912
|
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B
|
|
chr7_+_28914482
|
3.903
|
NM_001164655
|
9530053A07Rik
|
RIKEN cDNA 9530053A07 gene
|
|
chr10_+_58786043
|
3.871
|
NM_011030
|
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide
|
|
chr3_-_57379600
|
3.863
|
NM_133784
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr11_-_75236085
|
3.808
|
NM_011340
|
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1
|
|
chr11_+_57822347
|
3.785
|
NM_028451
|
Larp1
|
La ribonucleoprotein domain family, member 1
|
|
chrX_+_93291377
|
3.778
|
NM_010833
|
Msn
|
moesin
|
|
chr8_+_74134884
|
3.746
|
NM_146211
|
Glt25d1
|
glycosyltransferase 25 domain containing 1
|
|
chr5_+_97222355
|
3.731
|
NM_013470
|
Anxa3
|
annexin A3
|
|
chr2_-_58368568
|
3.728
|
|
Acvr1
|
activin A receptor, type 1
|
|
chr2_+_164595389
|
3.727
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr4_-_119331384
|
3.718
|
NM_008191
|
Guca2b
|
guanylate cyclase activator 2b (retina)
|
|
chr9_-_66891535
|
3.715
|
NM_001164252
NM_001164253
NM_001164254
NM_001164256
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr3_+_88433400
|
3.715
|
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr13_+_4058817
|
3.695
|
NM_134072
|
Akr1c14
|
aldo-keto reductase family 1, member C14
|
|
chr11_+_102110161
|
3.688
|
NM_153544
|
BC030867
|
cDNA sequence BC030867
|
|
chr3_+_122598292
|
3.677
|
NM_007980
|
Fabp2
|
fatty acid binding protein 2, intestinal
|
|
chrX_+_160811458
|
3.650
|
NM_010216
|
Figf
|
c-fos induced growth factor
|
|
chr1_-_121401236
|
3.624
|
NM_022327
|
Ralb
|
v-ral simian leukemia viral oncogene homolog B (ras related)
|
|
chr7_+_51066811
|
3.608
|
NM_011872
|
Klk7
|
kallikrein related-peptidase 7 (chymotryptic, stratum corneum)
|
|
chr13_+_21813912
|
3.604
|
NM_178200
|
Hist1h2bm
|
histone cluster 1, H2bm
|
|
chr19_+_58744854
|
3.600
|
NM_026925
|
Pnlip
|
pancreatic lipase
|
|
chr7_+_29649404
|
3.599
|
NM_008496
|
Lgals7
|
lectin, galactose binding, soluble 7
|
|
chr10_+_98570768
|
3.592
|
NM_015737
|
Galnt4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4
|
|
chr14_+_70499874
|
3.588
|
NM_021328
|
Bin3
|
bridging integrator 3
|
|
chr11_-_75236012
|
3.588
|
|
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1
|
|
chr4_+_133036046
|
3.574
|
NM_175307
|
Fam46b
|
family with sequence similarity 46, member B
|
|
chr7_+_29649467
|
3.574
|
|
Lgals7
|
lectin, galactose binding, soluble 7
|
|
chr12_+_32008372
|
3.564
|
|
Lamb1
|
laminin B1
|
|
chr8_+_97876182
|
3.538
|
NM_008609
|
Mmp15
|
matrix metallopeptidase 15
|
|
chr5_+_115373246
|
3.483
|
NM_145209
|
Oasl1
|
2'-5' oligoadenylate synthetase-like 1
|
|
chr16_-_85803359
|
3.483
|
NM_009621
|
Adamts1
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1
|
|
chr11_-_120434075
|
3.476
|
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide
|
|
chr13_+_22127689
|
3.439
|
NM_175665
|
Hist1h2bk
|
histone cluster 1, H2bk
|
|
chr7_-_29383178
|
3.438
|
|
Pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr13_-_55932777
|
3.423
|
NM_011097
|
Pitx1
|
paired-like homeodomain transcription factor 1
|
|
chr11_-_94938114
|
3.404
|
|
Itga3
|
integrin alpha 3
|
|
chr17_-_35096167
|
3.391
|
NM_010478
|
Hspa1b
Hspa1a
|
heat shock protein 1B
heat shock protein 1A
|
|
chr19_+_6276885
|
3.390
|
NM_010119
|
Ehd1
|
EH-domain containing 1
|
|
chr9_-_115152895
|
3.373
|
|
Stt3b
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae)
|
|
chr7_-_125635435
|
3.370
|
NM_001033380
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr3_-_100489321
|
3.370
|
|
Man1a2
|
mannosidase, alpha, class 1A, member 2
|
|
chr19_-_7070522
|
3.367
|
NM_026675
|
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22
|
|
chr1_+_74421706
|
3.358
|
NM_013612
|
Slc11a1
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1
|
|
chr11_+_48615792
|
3.353
|
|
Gnb2l1
|
guanine nucleotide binding protein (G protein), beta polypeptide 2 like 1
|
|
chr10_-_30338265
|
3.347
|
|
Hint3
|
histidine triad nucleotide binding protein 3
|
|
chr15_-_76036745
|
3.346
|
NM_001163540
NM_201389
|
Plec
|
plectin
|
|
chr15_-_50721552
|
3.337
|
NM_032000
|
Trps1
|
trichorhinophalangeal syndrome I (human)
|
|
chr10_+_110943762
|
3.336
|
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1
|
|
chr6_+_125299717
|
3.321
|
NM_011609
|
Tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a
|