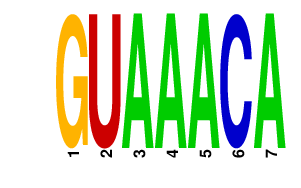
|
chr17_-_51965912
|
14.546
|
NM_001163631
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51972514
|
14.454
|
NM_001163632
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr2_-_18919637
|
13.640
|
NM_008845
|
Pip4k2a
|
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha
|
|
chr17_-_51971946
|
13.534
|
NM_009122
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr17_-_51951378
|
13.442
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr7_+_90015842
|
11.920
|
NM_175366
|
Mex3b
|
mex3 homolog B (C. elegans)
|
|
chr15_-_96472392
|
11.085
|
NM_134086
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr15_-_96473193
|
10.890
|
NM_001166458
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr11_-_116274264
|
10.401
|
NM_027258
|
Rnf157
|
ring finger protein 157
|
|
chr4_+_32504409
|
10.282
|
NM_001109661
|
Bach2
|
BTB and CNC homology 2
|
|
chr1_-_74590801
|
10.183
|
NM_176972
|
Usp37
|
ubiquitin specific peptidase 37
|
|
chr13_+_91600662
|
10.049
|
NM_024186
NM_024272
|
Ssbp2
|
single-stranded DNA binding protein 2
|
|
chr2_+_112106470
|
10.034
|
NM_133649
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chrX_-_54591905
|
9.712
|
NM_152801
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6
|
|
chr12_-_119539909
|
9.698
|
NM_001166385
NM_009239
|
Sp4
|
trans-acting transcription factor 4
|
|
chr9_+_74918979
|
9.614
|
NM_010864
|
Myo5a
|
myosin VA
|
|
chr9_+_57556346
|
9.332
|
NM_153799
|
Edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae)
|
|
chr7_+_95578782
|
9.145
|
NM_028238
|
Rab38
|
RAB38, member of RAS oncogene family
|
|
chr1_+_138027943
|
8.984
|
NM_001039472
|
Kif21b
|
kinesin family member 21B
|
|
chr8_-_112692071
|
8.955
|
NM_007586
|
Calb2
|
calbindin 2
|
|
chr1_+_132613903
|
8.871
|
NM_178691
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae)
|
|
chr7_+_133991066
|
8.684
|
NM_010069
|
Doc2a
|
double C2, alpha
|
|
chr8_+_73796512
|
8.532
|
NM_001142322
NM_001142323
NM_015742
|
Myo9b
|
myosin IXb
|
|
chrX_+_82819294
|
8.429
|
NM_025729
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3
|
|
chr5_+_72091188
|
8.336
|
NM_008069
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1
|
|
chr2_-_73367455
|
7.991
|
NM_153138
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr6_+_8459595
|
7.975
|
NM_133236
|
Glcci1
|
glucocorticoid induced transcript 1
|
|
chrX_-_11657678
|
7.882
|
NM_029510
NM_175044
NM_175045
NM_175046
|
Bcor
|
BCL6 interacting corepressor
|
|
chr3_+_103083215
|
7.880
|
NM_001079830
NM_053170
|
Trim33
|
tripartite motif-containing 33
|
|
chr2_+_112124767
|
7.600
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr8_+_116729508
|
7.595
|
NM_173016
|
Vat1l
|
vesicle amine transport protein 1 homolog-like (T. californica)
|
|
chr1_+_129999868
|
7.590
|
NM_181750
|
R3hdm1
|
R3H domain 1 (binds single-stranded nucleic acids)
|
|
chr15_-_98711692
|
7.532
|
NM_026967
|
Rhebl1
|
Ras homolog enriched in brain like 1
|
|
chr16_+_19760300
|
7.515
|
NM_001159407
NM_001159408
NM_054052
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
|
|
chr17_+_83750246
|
7.484
|
NM_001114361
NM_001114362
NM_199466
|
Eml4
|
echinoderm microtubule associated protein like 4
|
|
chr13_-_69878721
|
7.340
|
NM_001145162
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr11_+_104469313
|
7.291
|
NM_016780
|
Itgb3
|
integrin beta 3
|
|
chr3_+_55586431
|
7.184
|
NM_010750
|
Mab21l1
|
mab-21-like 1 (C. elegans)
|
|
chr11_-_79317583
|
7.144
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr1_+_155500166
|
7.137
|
NM_026380
|
Rgs8
|
regulator of G-protein signaling 8
|
|
chrX_+_133584130
|
7.137
|
NM_146261
|
Fam199x
|
family with sequence similarity 199, X-linked
|
|
chr16_-_46010510
|
7.024
|
NM_001134480
|
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2
|
|
chr6_+_118016385
|
7.017
|
NM_027526
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr4_-_82505534
|
6.902
|
NM_026647
|
Zdhhc21
|
zinc finger, DHHC domain containing 21
|
|
chr10_+_110601745
|
6.886
|
NM_001003717
NM_175489
|
Osbpl8
|
oxysterol binding protein-like 8
|
|
chr5_-_23536487
|
6.859
|
NM_053090
|
Fam126a
|
family with sequence similarity 126, member A
|
|
chr1_-_97564139
|
6.828
|
NM_001159745
NM_009183
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4
|
|
chr17_-_24826873
|
6.794
|
NM_011522
|
Syngr3
|
synaptogyrin 3
|
|
chr7_+_133415982
|
6.691
|
NM_145587
|
Sbk1
|
SH3-binding kinase 1
|
|
chr9_+_108728650
|
6.662
|
NM_080437
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr16_-_92697567
|
6.570
|
NM_001111023
NM_009821
|
Runx1
|
runt related transcription factor 1
|
|
chr1_-_90598670
|
6.359
|
NM_177305
|
Arl4c
|
ADP-ribosylation factor-like 4C
|
|
chr10_-_9620837
|
6.342
|
NM_001081344
|
Stxbp5
|
syntaxin binding protein 5 (tomosyn)
|
|
chr11_+_117515548
|
6.261
|
NM_198022
|
Tnrc6c
|
trinucleotide repeat containing 6C
|
|
chr7_+_107376116
|
6.204
|
NM_027629
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr10_+_82092582
|
6.190
|
NM_011561
NM_172552
|
Tdg
Gm9855
|
thymine DNA glycosylase
thymine DNA glycosylase pseudogene
|
|
chr5_-_123214252
|
6.078
|
NM_145358
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2, beta
|
|
chr8_-_70342105
|
6.074
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr17_+_25325344
|
6.059
|
NM_001197024
|
Unkl
|
unkempt-like (Drosophila)
|
|
chr19_+_57435498
|
6.014
|
NM_145505
|
Fam160b1
|
family with sequence similarity 160, member B1
|
|
chr10_-_118677059
|
6.009
|
NM_027994
|
Cand1
|
cullin associated and neddylation disassociated 1
|
|
chr8_-_30330079
|
6.002
|
NM_153135
|
Unc5d
|
unc-5 homolog D (C. elegans)
|
|
chr6_-_58857000
|
5.980
|
NM_021432
|
Nap1l5
|
nucleosome assembly protein 1-like 5
|
|
chrX_-_119510890
|
5.978
|
NM_138742
|
Nap1l3
|
nucleosome assembly protein 1-like 3
|
|
chrX_-_11737471
|
5.954
|
NM_001168321
|
Bcor
2900008C10Rik
|
BCL6 interacting corepressor
RIKEN cDNA 2900008C10 gene
|
|
chr5_-_68238654
|
5.942
|
NM_001038999
NM_009727
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr12_+_90191833
|
5.923
|
NM_172544
|
Nrxn3
|
neurexin III
|
|
chr15_+_30102306
|
5.899
|
NM_008729
|
Ctnnd2
|
catenin (cadherin associated protein), delta 2
|
|
chr19_+_38339167
|
5.891
|
NM_020278
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr10_+_39089594
|
5.880
|
NM_001122892
NM_001122893
NM_008054
|
Fyn
|
Fyn proto-oncogene
|
|
chr14_-_55735114
|
5.855
|
NM_177049
|
Jph4
|
junctophilin 4
|
|
chr17_+_25359526
|
5.848
|
NM_028789
|
Unkl
|
unkempt-like (Drosophila)
|
|
chr12_-_109241423
|
5.838
|
NM_001079883
NM_021399
|
Bcl11b
|
B-cell leukemia/lymphoma 11B
|
|
chr6_-_112897259
|
5.792
|
NM_080448
|
Srgap3
|
SLIT-ROBO Rho GTPase activating protein 3
|
|
chr12_+_12268901
|
5.763
|
NM_001146119
NM_029758
|
Fam49a
|
family with sequence similarity 49, member A
|
|
chr18_-_10610315
|
5.747
|
NM_001081222
|
Esco1
|
establishment of cohesion 1 homolog 1 (S. cerevisiae)
|
|
chr4_-_109959366
|
5.738
|
NM_001163399
NM_010488
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr4_-_110024513
|
5.733
|
NM_001163397
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr12_+_72116953
|
5.684
|
NM_001081195
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like)
|
|
chr6_+_86354212
|
5.681
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr3_+_136333733
|
5.600
|
NM_008913
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr3_+_94282752
|
5.512
|
NM_172434
|
Celf3
|
VCUGBP, Elav-like family member 3
|
|
chr2_-_66278870
|
5.488
|
NM_018733
|
Scn1a
|
sodium channel, voltage-gated, type I, alpha
|
|
chr6_-_113991945
|
5.479
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr10_-_80945445
|
5.474
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr2_+_4322020
|
5.471
|
NM_001177843
|
Frmd4a
|
FERM domain containing 4A
|
|
chr18_+_78135191
|
5.465
|
NM_001195633
|
5430411K18Rik
|
RIKEN cDNA 5430411K18 gene
|
|
chr9_-_24307580
|
5.440
|
NM_172920
|
Dpy19l1
|
dpy-19-like 1 (C. elegans)
|
|
chr7_-_134164627
|
5.414
|
NM_001102563
|
Prrt2
|
proline-rich transmembrane protein 2
|
|
chr17_+_36002537
|
5.388
|
NM_001146711
|
2310014H01Rik
|
RIKEN cDNA 2310014H01 gene
|
|
chr8_-_37312569
|
5.372
|
NM_001081150
|
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1
|
|
chr1_+_129670740
|
5.367
|
NM_028399
|
Ccnt2
|
cyclin T2
|
|
chr15_-_96472600
|
5.349
|
NM_001166456
|
Slc38a1
|
solute carrier family 38, member 1
|
|
chr2_-_66473008
|
5.315
|
NM_018852
|
Scn9a
|
sodium channel, voltage-gated, type IX, alpha
|
|
chr14_-_58508974
|
5.309
|
NM_029492
|
Zdhhc20
|
zinc finger, DHHC domain containing 20
|
|
chr8_-_9771020
|
5.308
|
NM_173446
|
Fam155a
|
family with sequence similarity 155, member A
|
|
chr16_+_87699143
|
5.263
|
NM_007520
|
Bach1
|
BTB and CNC homology 1
|
|
chr4_-_149050154
|
5.230
|
NM_001164051
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr8_+_128822478
|
5.229
|
NM_175434
|
Slc35f3
|
solute carrier family 35, member F3
|
|
chr2_+_4073908
|
5.226
|
NM_172475
|
Frmd4a
|
FERM domain containing 4A
|
|
chr3_-_152003317
|
5.224
|
NM_001162375
NM_174868
|
Fam73a
|
family with sequence similarity 73, member A
|
|
chr11_-_97605846
|
5.220
|
NM_054051
|
Pip4k2b
|
phosphatidylinositol-5-phosphate 4-kinase, type II, beta
|
|
chr16_-_31201251
|
5.216
|
NM_030138
|
Acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2
|
|
chr16_+_13358513
|
5.214
|
NM_153588
|
Mkl2
|
MKL/myocardin-like 2
|
|
chr2_+_65508501
|
5.193
|
NM_001099298
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1
|
|
chr4_+_21703416
|
5.172
|
NM_152825
|
Usp45
|
ubiquitin specific petidase 45
|
|
chr18_+_63868337
|
5.135
|
NM_001014981
|
Wdr7
|
WD repeat domain 7
|
|
chr7_+_130267398
|
5.134
|
NM_144925
|
Tnrc6a
|
trinucleotide repeat containing 6a
|
|
chr4_+_151712739
|
5.126
|
NM_001081376
NM_029216
|
Chd5
|
chromodomain helicase DNA binding protein 5
|
|
chr1_+_60466462
|
5.095
|
NM_001198570
NM_001198571
NM_198127
|
Abi2
|
abl-interactor 2
|
|
chr5_+_97426665
|
5.091
|
NM_080708
|
Bmp2k
|
BMP2 inducible kinase
|
|
chr11_-_69227176
|
5.050
|
NM_001017426
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B
|
|
chr10_-_62342761
|
5.033
|
NM_027384
|
Tet1
|
tet oncogene 1
|
|
chr16_+_52031661
|
4.991
|
NM_001033238
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr12_-_71448600
|
4.972
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr4_+_122860705
|
4.965
|
NM_174998
|
Hpcal4
|
hippocalcin-like 4
|
|
chr10_+_4424140
|
4.957
|
NM_001161822
NM_019958
|
Rgs17
|
regulator of G-protein signaling 17
|
|
chr2_-_74416925
|
4.956
|
NM_001110209
NM_027133
|
Lnp
|
limb and neural patterns
|
|
chr18_-_25912453
|
4.928
|
NM_001146292
NM_001146293
NM_001146294
NM_001146295
NM_001174074
NM_133195
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr11_-_17853850
|
4.881
|
NM_026576
|
Etaa1
|
Ewing's tumor-associated antigen 1
|
|
chr6_-_113841356
|
4.826
|
NM_001036684
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr9_+_26559146
|
4.824
|
NM_029792
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P)
|
|
chrX_+_23270202
|
4.809
|
NM_175180
|
Wdr44
|
WD repeat domain 44
|
|
chr13_-_100874021
|
4.804
|
NM_001081061
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB
|
|
chr7_-_36587975
|
4.761
|
NM_177739
|
Zfp507
E130304I02Rik
|
zinc finger protein 507
RIKEN cDNA E130304I02 gene
|
|
chr4_+_137150067
|
4.754
|
NM_130879
|
Usp48
|
ubiquitin specific peptidase 48
|
|
chr9_-_49607164
|
4.749
|
NM_001081445
NM_001113204
NM_010875
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr4_-_109960078
|
4.745
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr13_+_41701584
|
4.702
|
NM_001163572
|
Tmem170b
|
transmembrane protein 170B
|
|
chr1_-_156946739
|
4.699
|
NM_010500
|
Ier5
|
immediate early response 5
|
|
chr3_-_109946355
|
4.679
|
NM_001163348
NM_001163349
NM_001163350
NM_001163351
NM_030699
NM_133488
|
Ntng1
|
netrin G1
|
|
chr15_+_99984826
|
4.606
|
NM_172819
|
Dip2b
|
DIP2 disco-interacting protein 2 homolog B (Drosophila)
|
|
chr2_+_4480799
|
4.596
|
NM_001177844
|
Frmd4a
|
FERM domain containing 4A
|
|
chr3_-_19164956
|
4.564
|
NM_008802
|
Pde7a
|
phosphodiesterase 7A
|
|
chr8_+_41393330
|
4.562
|
NM_030110
|
Efha2
|
EF-hand domain family, member A2
|
|
chr1_-_162281701
|
4.558
|
NM_001038621
|
Rabgap1l
|
RAB GTPase activating protein 1-like
|
|
chr7_-_59117613
|
4.551
|
NM_001115087
|
Fancf
|
Fanconi anemia, complementation group F
|
|
chr11_-_107331865
|
4.550
|
NM_145823
|
Pitpnc1
|
phosphatidylinositol transfer protein, cytoplasmic 1
|
|
chr14_+_28435613
|
4.542
|
NM_177814
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr3_+_101814059
|
4.503
|
NM_178777
|
Nhlh2
|
nescient helix loop helix 2
|
|
chr1_-_108610861
|
4.486
|
NM_009741
NM_177410
|
Bcl2
|
B-cell leukemia/lymphoma 2
|
|
chr10_-_88720534
|
4.459
|
NM_178773
|
Ano4
|
anoctamin 4
|
|
chr7_-_140004777
|
4.454
|
NM_212473
|
Fam53b
|
family with sequence similarity 53, member B
|
|
chr17_+_70871444
|
4.433
|
NM_001128180
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr11_+_23156022
|
4.428
|
NM_001035226
NM_134014
|
Xpo1
|
exportin 1, CRM1 homolog (yeast)
|
|
chr1_-_78193381
|
4.421
|
NM_001159520
NM_008781
|
Pax3
|
paired box gene 3
|
|
chr17_+_49071906
|
4.390
|
NM_027452
|
Lrfn2
|
leucine rich repeat and fibronectin type III domain containing 2
|
|
chr2_+_112219335
|
4.389
|
NM_024254
|
2410042D21Rik
|
RIKEN cDNA 2410042D21 gene
|
|
chr10_+_19654195
|
4.364
|
NM_008580
|
Map3k5
|
mitogen-activated protein kinase kinase kinase 5
|
|
chr1_-_157820378
|
4.355
|
NM_001039184
|
Cep350
|
centrosomal protein 350
|
|
chr8_+_32222291
|
4.351
|
NM_175136
|
Rnf122
|
ring finger protein 122
|
|
chr10_-_110447050
|
4.311
|
NM_172554
|
Zdhhc17
|
zinc finger, DHHC domain containing 17
|
|
chr4_-_131817388
|
4.305
|
NM_001122992
NM_020273
|
Gmeb1
|
glucocorticoid modulatory element binding protein 1
|
|
chr2_+_164793487
|
4.290
|
NM_020333
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr4_-_91042997
|
4.258
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr12_+_33063653
|
4.238
|
NM_001162903
|
2010109K11Rik
|
RIKEN cDNA 2010109K11 gene
|
|
chr4_-_155366696
|
4.222
|
NM_080445
|
B3galt6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase, polypeptide 6
|
|
chr16_-_92826307
|
4.215
|
NM_001111021
NM_001111022
|
Runx1
|
runt related transcription factor 1
|
|
chr1_+_165860156
|
4.203
|
NM_028776
|
Scyl3
|
SCY1-like 3 (S. cerevisiae)
|
|
chr9_-_96531911
|
4.201
|
NM_053268
|
Rasa2
|
RAS p21 protein activator 2
|
|
chr17_+_74738195
|
4.196
|
NM_001162870
NM_016962
|
Spast
|
spastin
|
|
chr2_-_120229789
|
4.153
|
NM_001110496
NM_001110497
NM_173734
|
Tmem87a
|
transmembrane protein 87A
|
|
chr11_-_4018706
|
4.126
|
NM_144520
|
Sec14l2
|
SEC14-like 2 (S. cerevisiae)
|
|
chr12_+_50483869
|
4.124
|
NM_001160112
NM_008241
|
Foxg1
|
forkhead box G1
|
|
chr8_-_85856284
|
4.074
|
NM_001170691
NM_178736
|
Elmod2
|
ELMO domain containing 2
|
|
chr4_-_91066674
|
4.045
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr7_+_94732677
|
4.027
|
NM_001081414
NM_001143834
|
Grm5
|
glutamate receptor, metabotropic 5
|
|
chr10_-_13194201
|
4.020
|
NM_001195065
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr4_-_91038729
|
4.017
|
NM_010486
NM_207686
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr11_+_45869473
|
4.010
|
NM_009616
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr1_+_9838330
|
3.989
|
NM_001037759
|
Sgk3
|
serum/glucocorticoid regulated kinase 3
|
|
chrX_-_154036606
|
3.979
|
NM_172307
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2
|
|
chr9_+_72380046
|
3.977
|
NM_001033536
|
Rfx7
|
regulatory factor X, 7
|
|
chr17_-_80606644
|
3.947
|
NM_001195485
NM_001195486
NM_001195487
NM_146083
|
Srsf7
|
serine/arginine-rich splicing factor 7
|
|
chr7_-_139978601
|
3.937
|
NM_175268
|
Fam53b
|
family with sequence similarity 53, member B
|
|
chr17_+_36003527
|
3.933
|
NM_175242
|
2310014H01Rik
|
RIKEN cDNA 2310014H01 gene
|
|
chr3_-_78949796
|
3.931
|
NM_001099624
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2
|
|
chr15_-_78548542
|
3.919
|
NM_183141
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2
|
|
chr18_-_70212948
|
3.917
|
NM_030554
|
Rab27b
|
RAB27b, member RAS oncogene family
|
|
chr4_-_149075697
|
3.915
|
NM_001029837
|
Pik3cd
|
phosphatidylinositol 3-kinase catalytic delta polypeptide
|
|
chr7_-_64765360
|
3.894
|
NM_176942
|
Gabra5
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 5
|
|
chr5_+_19413335
|
3.878
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chrX_-_48559008
|
3.828
|
NM_134163
|
Mbnl3
|
muscleblind-like 3 (Drosophila)
|
|
chr10_+_84039622
|
3.828
|
NM_146008
|
Tcp11l2
|
t-complex 11 (mouse) like 2
|
|
chr2_+_119500777
|
3.822
|
NM_030112
|
Rtf1
|
Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr2_-_165710128
|
3.814
|
NM_027230
|
Zmynd8
|
zinc finger, MYND-type containing 8
|
|
chr8_-_107155195
|
3.806
|
NM_019662
|
Rrad
|
Ras-related associated with diabetes
|
|
chr18_-_43847367
|
3.799
|
NM_001163637
|
Jakmip2
|
janus kinase and microtubule interacting protein 2
|
|
chr2_-_91803660
|
3.793
|
NM_138306
|
Dgkz
|
diacylglycerol kinase zeta
|
|
chr8_+_108692750
|
3.773
|
NM_178798
|
Slc7a6
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 6
|
|
chr16_+_55973916
|
3.762
|
NM_173026
|
Zbtb11
|
zinc finger and BTB domain containing 11
|
|
chr9_-_75446888
|
3.751
|
NM_001038710
|
Tmod2
|
tropomodulin 2
|
|
chr13_+_99124196
|
3.748
|
NM_008242
|
Foxd1
|
forkhead box D1
|
|
chr9_-_75459125
|
3.747
|
NM_016711
|
Tmod2
|
tropomodulin 2
|
|
chr15_-_9070066
|
3.739
|
NM_013787
NM_145468
|
Skp2
|
S-phase kinase-associated protein 2 (p45)
|
|
chr4_-_120861138
|
3.714
|
NM_001081013
|
Rlf
|
rearranged L-myc fusion sequence
|
|
chr18_-_46358466
|
3.678
|
NM_001170855
|
Trim36
|
tripartite motif-containing 36
|
|
chr9_-_108092712
|
3.675
|
NM_007567
|
Bsn
|
bassoon
|
|
chr15_-_98701613
|
3.675
|
NM_001033276
|
Mll2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr2_-_139892477
|
3.658
|
NM_175225
NM_001159640
NM_001159641
|
Tasp1
|
taspase, threonine aspartase 1
|