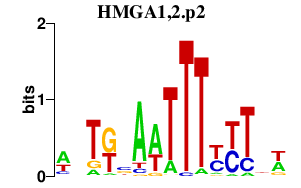
|
chr1_+_45402907
|
62.425
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_45368443
|
40.100
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_45390415
|
31.580
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_45401303
|
30.732
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr6_+_4490515
|
27.579
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr10_+_96944961
|
26.957
|
NM_007833
|
Dcn
|
decorin
|
|
chr6_+_4484651
|
21.361
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr5_-_17394564
|
19.771
|
|
Cd36
|
CD36 antigen
|
|
chr3_-_10208575
|
17.049
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr12_+_110783593
|
16.842
|
|
Meg3
|
maternally expressed 3
|
|
chr8_-_87297283
|
16.411
|
|
Nfix
|
nuclear factor I/X
|
|
chr1_+_45368382
|
15.688
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_-_172018982
|
14.341
|
NM_022563
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr1_-_66991625
|
14.132
|
NM_021285
|
Myl1
|
myosin, light polypeptide 1
|
|
chr15_-_3421486
|
13.937
|
NM_001048147
|
Ghr
|
growth hormone receptor
|
|
chr9_-_117161564
|
13.793
|
NM_001172122
NM_001172123
NM_001172124
NM_001172126
NM_178660
|
Rbms3
|
RNA binding motif, single stranded interacting protein
|
|
chr5_-_17355533
|
13.092
|
NM_001159556
NM_001159557
|
Cd36
|
CD36 antigen
|
|
chr6_+_4483741
|
12.861
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr12_+_110783786
|
12.771
|
|
Meg3
|
maternally expressed 3
|
|
chr1_-_66991580
|
12.702
|
|
Myl1
|
myosin, light polypeptide 1
|
|
chr2_+_131763454
|
12.671
|
|
Prnp
|
prion protein
|
|
chr14_-_104213981
|
12.607
|
|
|
|
|
chr6_+_34659440
|
12.558
|
NM_145575
|
Cald1
|
caldesmon 1
|
|
chr1_-_45559904
|
12.351
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr11_-_55208276
|
12.335
|
|
|
|
|
chr2_-_110203264
|
12.099
|
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr5_+_104864168
|
11.695
|
|
Spp1
|
secreted phosphoprotein 1
|
|
chr3_+_19857053
|
11.639
|
NM_001042611
NM_007752
|
Cp
|
ceruloplasmin
|
|
chr3_+_54165028
|
11.611
|
NM_001198765
NM_001198766
NM_015784
|
Postn
|
periostin, osteoblast specific factor
|
|
chr2_-_110203298
|
11.607
|
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr1_-_82232392
|
11.546
|
|
Irs1
|
insulin receptor substrate 1
|
|
chr15_+_3230053
|
11.301
|
|
|
|
|
chr1_-_142079842
|
11.255
|
NM_009888
|
Cfh
|
complement component factor h
|
|
chr3_-_129035406
|
11.156
|
NM_007934
|
Enpep
|
glutamyl aminopeptidase
|
|
chr17_-_14831095
|
10.961
|
|
Thbs2
|
thrombospondin 2
|
|
chr5_+_90889937
|
10.894
|
|
Alb
|
albumin
|
|
chr10_+_87322774
|
10.496
|
|
Igf1
|
insulin-like growth factor 1
|
|
chr7_-_51920870
|
10.467
|
NM_028021
|
Myh14
|
myosin, heavy polypeptide 14
|
|
chr10_+_87321789
|
10.338
|
NM_001111275
NM_010512
|
Igf1
|
insulin-like growth factor 1
|
|
chr12_+_101447413
|
10.325
|
|
Calm1
|
calmodulin 1
|
|
chr5_+_90889913
|
10.278
|
NM_009654
|
Alb
|
albumin
|
|
chr16_-_85173930
|
10.273
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr1_-_72905107
|
10.149
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr7_-_67149962
|
10.147
|
NM_013670
NM_033174
|
Snrpn
Snurf
|
small nuclear ribonucleoprotein N
SNRPN upstream reading frame
|
|
chr9_+_40590943
|
9.818
|
|
9030425E11Rik
|
RIKEN cDNA 9030425E11 gene
|
|
chr4_+_138012860
|
9.808
|
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr13_+_49703402
|
9.750
|
NM_008760
|
Ogn
|
osteoglycin
|
|
chr10_-_76172218
|
9.657
|
|
Col6a1
|
collagen, type VI, alpha 1
|
|
chr18_+_37906739
|
9.469
|
NM_033593
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr16_-_85173944
|
9.333
|
NM_001198823
NM_001198824
NM_001198825
NM_001198826
NM_007471
|
App
|
amyloid beta (A4) precursor protein
|
|
chr4_-_76240202
|
9.193
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chrX_-_91787372
|
9.028
|
NM_019791
|
Maged1
|
melanoma antigen, family D, 1
|
|
chr19_-_44470218
|
8.890
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr16_-_85173978
|
8.767
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr1_-_71686889
|
8.588
|
|
Fn1
|
fibronectin 1
|
|
chr16_-_85173918
|
8.526
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr3_+_79690856
|
8.518
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr2_-_58368568
|
8.517
|
|
Acvr1
|
activin A receptor, type 1
|
|
chr17_-_14831233
|
8.471
|
|
Thbs2
|
thrombospondin 2
|
|
chr10_+_69487700
|
8.321
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chr12_+_101447480
|
8.320
|
|
Calm1
|
calmodulin 1
|
|
chr14_+_27473359
|
8.201
|
|
Arf4
|
ADP-ribosylation factor 4
|
|
chr1_-_45560001
|
8.188
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr5_+_75548337
|
8.142
|
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr2_+_152512797
|
8.126
|
NM_025543
|
Mcts2
|
malignant T cell amplified sequence 2
|
|
chr2_+_136539143
|
8.053
|
NM_011428
|
Snap25
|
synaptosomal-associated protein 25
|
|
chr2_+_136539202
|
8.047
|
|
Snap25
|
synaptosomal-associated protein 25
|
|
chr10_+_24315247
|
7.972
|
NM_010217
|
Ctgf
|
connective tissue growth factor
|
|
chr8_-_11199802
|
7.961
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr10_+_87323855
|
7.958
|
NM_001111274
NM_001111276
NM_184052
|
Igf1
|
insulin-like growth factor 1
|
|
chr17_+_75578120
|
7.871
|
NM_206958
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr7_-_110976499
|
7.698
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr7_-_117509277
|
7.666
|
|
Sbf2
|
SET binding factor 2
|
|
chr7_-_148141821
|
7.637
|
NM_030694
|
Ifitm2
|
interferon induced transmembrane protein 2
|
|
chr3_-_144637974
|
7.611
|
NM_207208
|
Clca6
|
chloride channel calcium activated 6
|
|
chr3_-_113277601
|
7.492
|
NM_007446
|
Amy1
|
amylase 1, salivary
|
|
chr5_+_90890764
|
7.282
|
|
Alb
|
albumin
|
|
chr2_-_110203138
|
7.268
|
NM_026271
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr3_-_57105840
|
7.265
|
NM_008536
|
Tm4sf1
|
transmembrane 4 superfamily member 1
|
|
chr5_-_104543106
|
7.261
|
NM_010097
|
Sparcl1
|
SPARC-like 1
|
|
chr16_-_85173851
|
7.126
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chrX_+_135744582
|
7.121
|
NM_001164630
NM_175541
|
Mum1l1
|
melanoma associated antigen (mutated) 1-like 1
|
|
chr15_-_99609550
|
6.935
|
|
Lima1
|
LIM domain and actin binding 1
|
|
chr2_-_122420914
|
6.931
|
|
Gatm
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase)
|
|
chr2_-_140485267
|
6.911
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_-_122806122
|
6.906
|
NM_009779
|
C3ar1
|
complement component 3a receptor 1
|
|
chrX_-_132738383
|
6.906
|
NM_177919
|
Tceal5
|
transcription elongation factor A (SII)-like 5
|
|
chr9_-_15105864
|
6.845
|
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr6_+_56828743
|
6.842
|
|
Fkbp9
|
FK506 binding protein 9
|
|
chr17_-_14831262
|
6.817
|
NM_011581
|
Thbs2
|
thrombospondin 2
|
|
chr14_-_21277869
|
6.752
|
|
Anxa7
|
annexin A7
|
|
chr16_+_7409879
|
6.716
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr11_+_89891661
|
6.646
|
NM_026433
|
Tmem100
|
transmembrane protein 100
|
|
chr15_-_8660442
|
6.603
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr1_+_21230683
|
6.599
|
|
Gsta3
|
glutathione S-transferase, alpha 3
|
|
chr5_+_75548309
|
6.551
|
NM_011058
|
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide
|
|
chr1_-_66981900
|
6.539
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chr2_+_143768541
|
6.532
|
|
Dstn
|
destrin
|
|
chrX_+_93651684
|
6.496
|
NM_001159627
NM_181273
|
Heph
|
hephaestin
|
|
chr11_-_118163704
|
6.422
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr1_-_71642303
|
6.401
|
|
Fn1
|
fibronectin 1
|
|
chr2_+_145647868
|
6.304
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr5_-_104542641
|
6.235
|
|
Sparcl1
|
SPARC-like 1
|
|
chr15_-_8660729
|
6.199
|
NM_148938
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr7_+_121184667
|
6.191
|
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein
|
|
chr2_+_145648290
|
6.045
|
|
Rin2
|
Ras and Rab interactor 2
|
|
chr12_+_37968225
|
6.028
|
NM_178767
|
Tmem195
|
transmembrane protein 195
|
|
chr9_-_15105965
|
5.972
|
NM_133732
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr16_+_34921598
|
5.925
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr15_-_8660462
|
5.919
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr3_-_122241909
|
5.894
|
|
Fnbp1l
|
formin binding protein 1-like
|
|
chr6_-_36761360
|
5.844
|
NM_008973
|
Ptn
|
pleiotrophin
|
|
chr14_+_63756868
|
5.741
|
|
Ctsb
|
cathepsin B
|
|
chr19_+_23239333
|
5.739
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr16_-_84955086
|
5.723
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr2_-_84490666
|
5.705
|
NM_001085453
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chr9_-_49311823
|
5.696
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr8_-_67071497
|
5.655
|
|
|
|
|
chr5_-_67009869
|
5.637
|
NM_009686
|
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr15_+_3220766
|
5.603
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr17_-_56419388
|
5.516
|
|
Plin3
|
perilipin 3
|
|
chr2_+_128604653
|
5.499
|
|
Mertk
|
c-mer proto-oncogene tyrosine kinase
|
|
chr15_-_96861590
|
5.489
|
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr9_+_43877511
|
5.485
|
NM_016808
|
Usp2
|
ubiquitin specific peptidase 2
|
|
chr17_-_14831058
|
5.468
|
|
Thbs2
|
thrombospondin 2
|
|
chr10_-_60227439
|
5.424
|
|
Unc5b
|
unc-5 homolog B (C. elegans)
|
|
chr17_+_43526389
|
5.423
|
NM_001081178
|
Gpr116
|
G protein-coupled receptor 116
|
|
chr1_+_151947225
|
5.406
|
NM_011198
|
Ptgs2
|
prostaglandin-endoperoxide synthase 2
|
|
chr8_-_49760037
|
5.371
|
NM_001145937
NM_011857
|
Odz3
|
odd Oz/ten-m homolog 3 (Drosophila)
|
|
chr2_+_72244049
|
5.311
|
|
B230120H23Rik
|
RIKEN cDNA B230120H23 gene
|
|
chr3_+_14863507
|
5.301
|
NM_007606
|
Car3
|
carbonic anhydrase 3
|
|
chr16_+_42955690
|
5.270
|
|
BC002163
|
NADH dehydrogenase Fe-S protein 5 pseudogene
|
|
chr4_-_142889372
|
5.266
|
NM_010329
|
Pdpn
|
podoplanin
|
|
chr9_+_110255447
|
5.265
|
|
|
|
|
chr18_+_20823908
|
5.259
|
|
Ttr
|
transthyretin
|
|
chr7_-_52230179
|
5.250
|
|
Cpt1c
|
carnitine palmitoyltransferase 1c
|
|
chr1_-_95518831
|
5.221
|
|
Stk25
|
serine/threonine kinase 25 (yeast)
|
|
chr7_+_27904997
|
5.218
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr10_+_97028456
|
5.200
|
|
Lum
|
lumican
|
|
chr19_+_23240506
|
5.199
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr15_-_8660479
|
5.171
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr12_+_110884340
|
5.142
|
|
Rian
|
RNA imprinted and accumulated in nucleus
|
|
chr2_+_71892681
|
5.134
|
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4
|
|
chr12_+_32008372
|
5.127
|
|
Lamb1
|
laminin B1
|
|
chr4_-_60594469
|
5.078
|
|
Mup1
|
major urinary protein 1
|
|
chrX_+_20127488
|
5.067
|
|
Rgn
|
regucalcin
|
|
chr10_+_56107352
|
5.065
|
|
Gja1
|
gap junction protein, alpha 1
|
|
chr4_+_41709182
|
5.027
|
NM_010549
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1
|
|
chr4_-_61179982
|
5.027
|
|
Mup1
|
major urinary protein 1
|
|
chr10_-_83989221
|
4.998
|
|
Ckap4
|
cytoskeleton-associated protein 4
|
|
chr18_+_37896589
|
4.987
|
NM_033592
|
Pcdhga9
|
protocadherin gamma subfamily A, 9
|
|
chr3_-_97628441
|
4.958
|
NM_001110163
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin)
|
|
chr5_-_104542598
|
4.943
|
|
Sparcl1
|
SPARC-like 1
|
|
chr18_+_37906668
|
4.930
|
|
Pcdhga10
|
protocadherin gamma subfamily A, 10
|
|
chr5_-_100812885
|
4.903
|
|
Sec31a
|
Sec31 homolog A (S. cerevisiae)
|
|
chr14_-_66572577
|
4.884
|
NM_172604
|
Scara3
|
scavenger receptor class A, member 3
|
|
chr1_-_72906185
|
4.859
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr5_+_115916274
|
4.857
|
NM_011107
|
Pla2g1b
|
phospholipase A2, group IB, pancreas
|
|
chr14_+_51710846
|
4.841
|
|
Rnase4
|
ribonuclease, RNase A family 4
|
|
chr1_+_59543450
|
4.836
|
|
|
|
|
chr3_-_10208480
|
4.825
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr12_+_110779204
|
4.823
|
|
Meg3
|
maternally expressed 3
|
|
chr5_-_104542751
|
4.814
|
|
Sparcl1
|
SPARC-like 1
|
|
chrX_+_160811458
|
4.808
|
NM_010216
|
Figf
|
c-fos induced growth factor
|
|
chr5_-_67009992
|
4.748
|
|
Apbb2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr5_-_17341534
|
4.689
|
|
Cd36
|
CD36 antigen
|
|
chr6_+_22825464
|
4.689
|
NM_001081306
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1
|
|
chr4_-_60594496
|
4.667
|
|
Mup1
|
major urinary protein 1
|
|
chr4_-_98049833
|
4.645
|
NM_053157
|
Tm2d1
|
TM2 domain containing 1
|
|
chr5_-_17394776
|
4.638
|
NM_001159558
|
Cd36
|
CD36 antigen
|
|
chr19_-_21467250
|
4.620
|
|
Gda
|
guanine deaminase
|
|
chr14_+_51627425
|
4.611
|
NM_001162863
|
Rnase10
|
ribonuclease, RNase A family, 10 (non-active)
|
|
chr17_-_56419161
|
4.567
|
|
Plin3
|
perilipin 3
|
|
chr7_-_110976438
|
4.563
|
|
Hbb-b1
|
hemoglobin, beta adult major chain
|
|
chr17_+_75784448
|
4.529
|
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr10_+_97028134
|
4.529
|
NM_008524
|
Lum
|
lumican
|
|
chr11_+_29640992
|
4.522
|
|
Rtn4
|
reticulon 4
|
|
chr3_-_148617598
|
4.519
|
NM_001081298
|
Lphn2
|
latrophilin 2
|
|
chr13_+_49704471
|
4.510
|
|
Ogn
|
osteoglycin
|
|
chr2_-_148701203
|
4.510
|
NM_009976
|
Cst3
|
cystatin C
|
|
chr7_-_110976314
|
4.509
|
|
Hbb-b1
Hbb-b2
|
hemoglobin, beta adult major chain
hemoglobin, beta adult minor chain
|
|
chr5_-_104421860
|
4.505
|
|
Hsd17b11
|
hydroxysteroid (17-beta) dehydrogenase 11
|
|
chr6_-_122806042
|
4.483
|
|
C3ar1
|
complement component 3a receptor 1
|
|
chr15_+_91991704
|
4.482
|
NM_001159647
|
Cntn1
|
contactin 1
|
|
chr4_-_142889312
|
4.472
|
|
Pdpn
|
podoplanin
|
|
chr10_+_96945057
|
4.461
|
|
Dcn
|
decorin
|
|
chr17_-_12888210
|
4.448
|
|
Igf2r
|
insulin-like growth factor 2 receptor
|
|
chr13_+_3553690
|
4.432
|
|
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2
|
|
chr14_+_76354807
|
4.425
|
NM_026214
|
Kctd4
|
potassium channel tetramerisation domain containing 4
|
|
chr18_+_37921454
|
4.403
|
NM_033580
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8
|
|
chr10_+_68996455
|
4.380
|
NM_146005
NM_170688
NM_170689
NM_170690
NM_170728
NM_170729
NM_170730
|
Ank3
|
ankyrin 3, epithelial
|
|
chr7_+_19661548
|
4.354
|
NM_010058
|
Dmwd
|
dystrophia myotonica-containing WD repeat motif
|
|
chr19_-_34329804
|
4.333
|
NM_007392
|
Acta2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr12_+_110806715
|
4.327
|
|
Meg3
|
maternally expressed 3
|
|
chr3_+_59835700
|
4.311
|
NM_023383
|
Aadac
|
arylacetamide deacetylase (esterase)
|
|
chr9_+_92502908
|
4.305
|
|
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr6_-_54543121
|
4.302
|
NM_153573
|
Fkbp14
|
FK506 binding protein 14
|
|
chr16_+_37884098
|
4.302
|
|
Lrrc58
|
leucine rich repeat containing 58
|
|
chr5_+_143734622
|
4.301
|
|
|
|
|
chr1_-_141982814
|
4.279
|
|
Cfh
|
complement component factor h
|