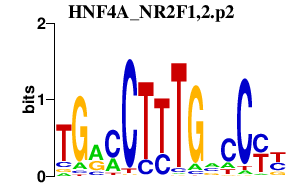
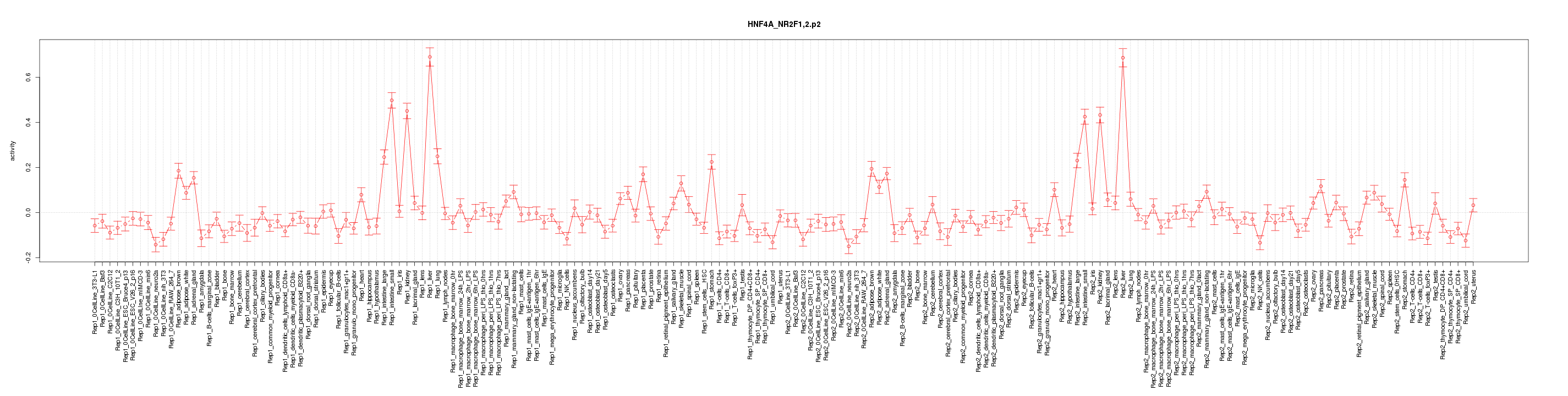
|
chr16_+_23146608
|
66.772
|
NM_009605
|
Adipoq
|
adiponectin, C1Q and collagen domain containing
|
|
chr16_+_23146631
|
65.661
|
|
Adipoq
|
adiponectin, C1Q and collagen domain containing
|
|
chr16_+_5007426
|
62.376
|
|
Gm5480
|
predicted gene 5480
|
|
chr2_+_172978566
|
58.944
|
NM_011044
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic
|
|
chr3_+_122598292
|
52.638
|
NM_007980
|
Fabp2
|
fatty acid binding protein 2, intestinal
|
|
chr14_-_56377206
|
49.852
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr14_-_56377231
|
49.796
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr14_-_56377278
|
49.458
|
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr14_-_56377260
|
49.111
|
NM_009894
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B
|
|
chr11_-_73137838
|
44.509
|
|
Aspa
|
aspartoacylase
|
|
chr17_-_43639675
|
43.356
|
NM_008585
|
Mep1a
|
meprin 1 alpha
|
|
chr9_+_46036703
|
42.983
|
NM_009692
|
Apoa1
|
apolipoprotein A-I
|
|
chr18_+_21230844
|
40.793
|
NM_008586
|
Mep1b
|
meprin 1 beta
|
|
chr11_-_77707282
|
39.019
|
NM_008952
|
Pipox
|
pipecolic acid oxidase
|
|
chr12_+_37968225
|
38.968
|
NM_178767
|
Tmem195
|
transmembrane protein 195
|
|
chr9_+_46036788
|
38.562
|
|
Apoa1
|
apolipoprotein A-I
|
|
chr4_-_129851289
|
37.499
|
|
Tinagl1
|
tubulointerstitial nephritis antigen-like 1
|
|
chr9_+_46036799
|
36.528
|
|
Apoa1
|
apolipoprotein A-I
|
|
chr1_+_74455956
|
35.825
|
NM_009509
|
Vil1
|
villin 1
|
|
chr7_+_150659707
|
34.871
|
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18
|
|
chr7_+_150659625
|
33.958
|
NM_008767
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18
|
|
chr15_+_82282806
|
33.088
|
NM_010006
|
Cyp2d9
|
cytochrome P450, family 2, subfamily d, polypeptide 9
|
|
chr6_+_71149869
|
32.949
|
NM_017399
|
Fabp1
|
fatty acid binding protein 1, liver
|
|
chr4_-_61179982
|
31.676
|
|
Mup1
|
major urinary protein 1
|
|
chr9_-_15105965
|
31.620
|
NM_133732
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chrX_+_136343083
|
31.156
|
|
Cldn2
|
claudin 2
|
|
chr3_+_94736993
|
30.992
|
NM_009150
|
Selenbp1
|
selenium binding protein 1
|
|
chr10_-_88969371
|
30.980
|
NM_001163700
NM_009108
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4
|
|
chr19_+_23239333
|
30.774
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr1_+_162917586
|
30.753
|
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr4_-_60235470
|
30.511
|
NM_001134676
|
Mup8
|
major urinary protein 8
|
|
chr17_-_34999458
|
30.151
|
NM_001142706
NM_008198
|
Cfb
|
complement factor B
|
|
chr1_+_162917640
|
30.101
|
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1
|
|
chr3_+_107680147
|
30.001
|
NM_133867
|
Eps8l3
|
EPS8-like 3
|
|
chr3_+_94736974
|
29.665
|
|
Selenbp1
|
selenium binding protein 1
|
|
chr9_-_15105864
|
29.583
|
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr10_-_128397928
|
28.794
|
NM_027853
|
Mettl7b
|
methyltransferase like 7B
|
|
chr17_+_24879324
|
28.480
|
|
Sepx1
|
selenoprotein X 1
|
|
chr11_-_73138102
|
28.433
|
NM_023113
|
Aspa
|
aspartoacylase
|
|
chr8_+_107450138
|
27.801
|
NM_001163756
NM_172759
|
Ces2e
|
carboxylesterase 2E
|
|
chr7_-_106844083
|
27.722
|
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1
|
|
chr17_-_34999068
|
27.625
|
|
Cfb
|
complement factor B
|
|
chr7_-_31840647
|
27.569
|
NM_019503
NM_052992
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr11_+_32183671
|
27.411
|
NM_001083955
NM_008218
|
Hba-a2
Hba-a1
|
hemoglobin alpha, adult chain 2
hemoglobin alpha, adult chain 1
|
|
chr15_+_3220766
|
27.232
|
NM_001042613
NM_001042614
NM_009155
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr11_+_32196488
|
27.141
|
NM_001083955
NM_008218
|
Hba-a2
Hba-a1
|
hemoglobin alpha, adult chain 2
hemoglobin alpha, adult chain 1
|
|
chr15_+_3220999
|
27.002
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr11_+_58192533
|
26.968
|
NM_001083884
NM_027339
|
2210415F13Rik
|
RIKEN cDNA 2210415F13 gene
|
|
chr15_-_3421486
|
26.340
|
NM_001048147
|
Ghr
|
growth hormone receptor
|
|
chr3_+_94497485
|
26.280
|
NM_019414
|
Selenbp2
|
selenium binding protein 2
|
|
chr13_-_41923709
|
25.624
|
|
9530008L14Rik
|
RIKEN cDNA 9530008L14 gene
|
|
chr15_+_3220994
|
25.542
|
|
Sepp1
|
selenoprotein P, plasma, 1
|
|
chr19_+_4594318
|
25.163
|
NM_008797
|
Pcx
|
pyruvate carboxylase
|
|
chr17_-_35018943
|
24.807
|
|
C2
|
complement component 2 (within H-2S)
|
|
chr1_-_45982242
|
24.723
|
NM_016917
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr6_+_72306146
|
23.824
|
|
Tmem150a
|
transmembrane protein 150A
|
|
chr3_-_129035406
|
23.795
|
NM_007934
|
Enpep
|
glutamyl aminopeptidase
|
|
chr11_-_3831515
|
23.745
|
|
Tcn2
|
transcobalamin 2
|
|
chr8_+_58773321
|
23.658
|
NM_008278
|
Hpgd
|
hydroxyprostaglandin dehydrogenase 15 (NAD)
|
|
chr17_-_34960292
|
23.460
|
NM_011413
|
C4a
|
complement component 4A (Rodgers blood group)
|
|
chr19_-_44482138
|
23.451
|
NM_009127
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr9_-_46043339
|
23.110
|
NM_023114
|
Apoc3
|
apolipoprotein C-III
|
|
chr19_-_44482033
|
23.064
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr7_-_31840594
|
22.999
|
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chr17_-_34880784
|
22.830
|
NM_009780
|
C4b
|
complement component 4B (Childo blood group)
|
|
chr7_+_50699457
|
22.795
|
NM_026695
|
Etfb
|
electron transferring flavoprotein, beta polypeptide
|
|
chr1_-_122017495
|
22.656
|
NM_001037999
|
Dbi
|
diazepam binding inhibitor
|
|
chr11_-_83406475
|
22.434
|
NM_009139
|
Ccl6
|
chemokine (C-C motif) ligand 6
|
|
chr6_-_124492297
|
22.302
|
NM_144938
NM_001097617
|
C1s
|
complement component 1, s subcomponent
|
|
chr15_+_101933364
|
22.278
|
NM_153533
|
Tenc1
|
tensin like C1 domain-containing phosphatase
|
|
chr19_+_46206383
|
21.904
|
NM_007703
|
Elovl3
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 3
|
|
chr16_+_8513513
|
21.758
|
NM_001170978
NM_172961
|
Abat
|
4-aminobutyrate aminotransferase
|
|
chr19_-_44481898
|
21.509
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr17_-_57354126
|
21.442
|
|
C3
|
complement component 3
|
|
chr6_-_128476708
|
21.068
|
NM_007376
|
Pzp
|
pregnancy zone protein
|
|
chr6_+_90424727
|
20.978
|
|
Klf15
|
Kruppel-like factor 15
|
|
chr6_+_138089041
|
20.190
|
NM_019946
|
Mgst1
|
microsomal glutathione S-transferase 1
|
|
chr7_+_148106800
|
20.130
|
NM_001081389
|
Nlrp6
|
NLR family, pyrin domain containing 6
|
|
chr13_-_94407590
|
20.047
|
NM_016668
|
Bhmt
|
betaine-homocysteine methyltransferase
|
|
chr8_+_11443165
|
19.567
|
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr19_-_44471675
|
19.474
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr2_+_122090555
|
19.474
|
|
Sord
|
sorbitol dehydrogenase
|
|
chr3_+_98084352
|
19.155
|
NM_008256
|
Hmgcs2
|
3-hydroxy-3-methylglutaryl-Coenzyme A synthase 2
|
|
chr1_+_159456274
|
19.023
|
NM_001159986
|
Sec16b
|
SEC16 homolog B (S. cerevisiae)
|
|
chr1_-_157659936
|
18.936
|
NM_001024945
NM_023268
|
Qsox1
|
quiescin Q6 sulfhydryl oxidase 1
|
|
chr19_-_39961258
|
18.887
|
NM_001104525
|
Cyp2c69
|
cytochrome P450, family 2, subfamily c, polypeptide 69
|
|
chr3_+_90346956
|
18.867
|
|
S100a16
|
S100 calcium binding protein A16
|
|
chr3_+_59835700
|
18.816
|
NM_023383
|
Aadac
|
arylacetamide deacetylase (esterase)
|
|
chr9_-_21940154
|
18.749
|
NM_001102405
|
Acp5
|
acid phosphatase 5, tartrate resistant
|
|
chr19_-_39887217
|
18.588
|
NM_010004
|
Cyp2c40
|
cytochrome P450, family 2, subfamily c, polypeptide 40
|
|
chr10_-_80555271
|
18.573
|
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3
|
|
chr16_+_33794452
|
18.431
|
|
Muc13
|
mucin 13, epithelial transmembrane
|
|
chr12_-_104976382
|
18.427
|
NM_009244
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B
|
|
chr4_-_61335031
|
18.302
|
|
Mup10
LOC100048884
Mup19
Mup1
Mup2
|
major urinary protein 10
novel member of the major urinary protein (Mup) gene family
major urinary protein 19
major urinary protein 1
major urinary protein 2
|
|
chr11_+_101229029
|
18.296
|
NM_008061
|
G6pc
|
glucose-6-phosphatase, catalytic
|
|
chr2_-_110145819
|
18.257
|
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase)
|
|
chr11_-_3831963
|
18.112
|
NM_001130459
NM_015749
NM_001130458
|
Tcn2
|
transcobalamin 2
|
|
chr5_-_136220001
|
18.110
|
NM_172541
|
Tmem120a
|
transmembrane protein 120A
|
|
chr5_-_146821173
|
17.977
|
NM_019792
|
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25
|
|
chr7_-_52830518
|
17.930
|
|
0610005C13Rik
|
RIKEN cDNA 0610005C13 gene
|
|
chr5_+_146890818
|
17.832
|
NM_001105160
|
Cyp3a59
|
cytochrome P450, subfamily 3A, polypeptide 59
|
|
chr11_+_98308147
|
17.776
|
NM_010346
|
Grb7
|
growth factor receptor bound protein 7
|
|
chr7_-_26181005
|
17.695
|
NM_010719
|
Lipe
|
lipase, hormone sensitive
|
|
chr17_-_35019039
|
17.682
|
NM_013484
|
C2
|
complement component 2 (within H-2S)
|
|
chr19_-_44472087
|
17.227
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr3_-_89084872
|
17.225
|
NM_001162425
|
Efna1
|
ephrin A1
|
|
chr10_-_80555389
|
17.138
|
|
Creb3l3
|
cAMP responsive element binding protein 3-like 3
|
|
chr11_+_98308333
|
17.119
|
|
Grb7
|
growth factor receptor bound protein 7
|
|
chr7_+_27620354
|
17.090
|
NM_007812
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr5_-_115569276
|
17.088
|
NM_007383
|
Acads
|
acyl-Coenzyme A dehydrogenase, short chain
|
|
chr19_+_39361574
|
17.077
|
NM_007815
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29
|
|
chr2_+_121183509
|
16.905
|
|
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous
|
|
chr15_+_98929891
|
16.873
|
NM_176835
|
Dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22
|
|
chr3_+_118265066
|
16.804
|
NM_170778
|
Dpyd
|
dihydropyrimidine dehydrogenase
|
|
chr17_-_46575402
|
16.764
|
NM_144856
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7
|
|
chr19_+_39081508
|
16.751
|
NM_028089
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55
|
|
chr1_+_173155414
|
16.742
|
|
Apoa2
|
apolipoprotein A-II
|
|
chr6_+_48791616
|
16.697
|
NM_001098271
|
Tmem176a
|
transmembrane protein 176A
|
|
chr7_+_51480806
|
16.666
|
NM_010639
|
Klk1
|
kallikrein 1
|
|
chr10_+_127213437
|
16.596
|
NM_153133
|
Rdh9
|
retinol dehydrogenase 9
|
|
chr16_+_31422377
|
16.543
|
NM_175177
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1
|
|
chr3_+_90345082
|
16.522
|
NM_026416
|
S100a16
|
S100 calcium binding protein A16
|
|
chr15_-_82237600
|
16.432
|
NM_010005
|
Cyp2d10
|
cytochrome P450, family 2, subfamily d, polypeptide 10
|
|
chr19_-_8206403
|
16.302
|
NM_001013820
|
Gm5631
|
predicted gene 5631
|
|
chr10_-_6806261
|
16.058
|
NM_027391
|
Iyd
|
iodotyrosine deiodinase
|
|
chr13_-_94407455
|
16.041
|
|
Bhmt
|
betaine-homocysteine methyltransferase
|
|
chr7_+_27620380
|
15.961
|
|
Cyp2a4
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 4
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr17_-_35268696
|
15.949
|
NM_018816
|
Apom
|
apolipoprotein M
|
|
chr19_-_8479532
|
15.937
|
NM_177002
|
C730048C13Rik
|
RIKEN cDNA C730048C13 gene
|
|
chr2_+_169459145
|
15.889
|
NM_080455
|
Tshz2
|
teashirt zinc finger family member 2
|
|
chr9_-_121825407
|
15.790
|
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1
|
|
chr2_-_13413502
|
15.704
|
NM_001081084
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor)
|
|
chr19_+_12707680
|
15.631
|
NM_145935
|
Glyat
|
glycine-N-acyltransferase
|
|
chr10_+_58866370
|
15.565
|
NM_023530
|
Pla2g12b
|
phospholipase A2, group XIIB
|
|
chr17_-_31266539
|
15.474
|
NM_011575
|
Tff3
|
trefoil factor 3, intestinal
|
|
chr7_+_29618812
|
15.471
|
NM_010706
|
Lgals4
Lgals6
|
lectin, galactose binding, soluble 4
lectin, galactose binding, soluble 6
|
|
chr16_-_21787906
|
15.280
|
NM_023737
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase
|
|
chr7_+_27904997
|
15.279
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr7_+_127317371
|
15.223
|
NM_028085
|
Anks4b
|
ankyrin repeat and sterile alpha motif domain containing 4B
|
|
chr9_-_121825422
|
15.132
|
NM_010012
|
Cyp8b1
|
cytochrome P450, family 8, subfamily b, polypeptide 1
|
|
chr17_-_46575337
|
15.125
|
|
Slc22a7
|
solute carrier family 22 (organic anion transporter), member 7
|
|
chr2_-_110145870
|
15.014
|
NM_130452
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase)
|
|
chr10_+_81022283
|
14.921
|
|
Aes
|
amino-terminal enhancer of split
|
|
chr6_+_124613114
|
14.884
|
NM_145130
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3
|
|
chr1_+_173155184
|
14.862
|
NM_013474
|
Apoa2
|
apolipoprotein A-II
|
|
chr5_-_87964170
|
14.857
|
|
Sult1b1
|
sulfotransferase family 1B, member 1
|
|
chr7_+_51720804
|
14.779
|
NM_026690
|
Aspdh
|
aspartate dehydrogenase domain containing
|
|
chrX_+_9829439
|
14.773
|
NM_008769
|
Otc
|
ornithine transcarbamylase
|
|
chrX_+_20126893
|
14.771
|
NM_009060
|
Rgn
|
regucalcin
|
|
chr11_-_100007218
|
14.727
|
NM_008471
|
Krt19
|
keratin 19
|
|
chr9_+_62705890
|
14.714
|
NM_001102468
NM_138304
|
Calml4
|
calmodulin-like 4
|
|
chr15_+_101980990
|
14.626
|
NM_146064
|
Soat2
|
sterol O-acyltransferase 2
|
|
chr3_-_137794277
|
14.567
|
NM_008642
|
Mttp
|
microsomal triglyceride transfer protein
|
|
chr5_-_104406333
|
14.409
|
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13
|
|
chr7_+_147279293
|
14.326
|
NM_009475
|
Prap1
|
proline-rich acidic protein 1
|
|
chr10_-_25255955
|
14.307
|
NM_001195596
|
2010003K15Rik
|
RIKEN cDNA 2010003K15 gene
|
|
chr14_+_32031041
|
14.297
|
NM_001025379
|
Sema3g
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G
|
|
chr15_-_96850752
|
14.264
|
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr16_+_33794122
|
14.239
|
NM_010739
|
Muc13
|
mucin 13, epithelial transmembrane
|
|
chr19_-_39815529
|
14.139
|
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68
|
|
chr18_+_85020800
|
14.135
|
NM_025797
|
Cyb5
|
cytochrome b-5
|
|
chr16_-_21787868
|
14.089
|
|
Ehhadh
|
enoyl-Coenzyme A, hydratase/3-hydroxyacyl Coenzyme A dehydrogenase
|
|
chr1_-_88998180
|
14.052
|
NM_001081082
|
Alpi
|
alkaline phosphatase, intestinal
|
|
chr15_+_6395353
|
14.047
|
|
C9
|
complement component 9
|
|
chr10_+_81022229
|
14.037
|
|
Aes
|
amino-terminal enhancer of split
|
|
chr4_+_97444312
|
14.029
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr12_-_86715324
|
13.980
|
|
Tmed10
|
transmembrane emp24-like trafficking protein 10 (yeast)
|
|
chr7_-_142720274
|
13.944
|
NM_178669
|
Clrn3
|
clarin 3
|
|
chr18_+_85020742
|
13.891
|
|
Cyb5
|
cytochrome b-5
|
|
chr7_-_26180629
|
13.812
|
|
Lipe
|
lipase, hormone sensitive
|
|
chr3_-_79432487
|
13.751
|
NM_025794
|
Etfdh
|
electron transferring flavoprotein, dehydrogenase
|
|
chr3_-_121518046
|
13.696
|
NM_008991
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3
|
|
chr10_+_81022304
|
13.675
|
NM_010347
|
Aes
|
amino-terminal enhancer of split
|
|
chr9_-_103119803
|
13.663
|
|
Trf
|
transferrin
|
|
chrX_+_93650774
|
13.635
|
NM_001159628
NM_010417
|
Heph
|
hephaestin
|
|
chr9_+_46048812
|
13.617
|
NM_007468
|
Apoa4
|
apolipoprotein A-IV
|
|
chr8_-_95753501
|
13.596
|
NM_133660
|
Ces1e
|
carboxylesterase 1E
|
|
chr12_+_7984451
|
13.530
|
NM_009693
|
Apob
|
apolipoprotein B
|
|
chr3_+_137949734
|
13.478
|
|
Adh1
|
alcohol dehydrogenase 1 (class I)
|
|
chr17_+_57201623
|
13.475
|
NM_177638
|
Crb3
|
crumbs homolog 3 (Drosophila)
|
|
chr4_-_137923869
|
13.423
|
NM_028176
|
Cda
|
cytidine deaminase
|
|
chr17_-_26005645
|
13.361
|
|
Fam195a
|
family with sequence similarity 195, member A
|
|
chr2_-_25356025
|
13.236
|
NM_027062
|
C8g
|
complement component 8, gamma polypeptide
|
|
chr9_+_107479247
|
13.229
|
NM_008317
|
Hyal1
|
hyaluronoglucosaminidase 1
|
|
chr18_+_20716598
|
13.215
|
NM_007883
|
Dsg2
|
desmoglein 2
|
|
chr11_-_4604308
|
13.147
|
NM_197979
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr9_+_46048993
|
13.056
|
|
Apoa4
|
apolipoprotein A-IV
|
|
chr16_-_20716708
|
13.040
|
NM_009900
|
Clcn2
|
chloride channel 2
|
|
chr8_-_13200534
|
13.008
|
NM_025768
|
Grtp1
|
GH regulated TBC protein 1
|
|
chr12_-_85741801
|
13.001
|
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5
|
|
chr11_-_53236630
|
12.937
|
NM_153069
|
Leap2
|
liver-expressed antimicrobial peptide 2
|
|
chr8_+_46035561
|
12.908
|
NM_001081286
|
Fat1
|
FAT tumor suppressor homolog 1 (Drosophila)
|
|
chr18_+_74893369
|
12.908
|
|
Myo5b
|
myosin VB
|
|
chr7_-_52359079
|
12.905
|
NM_010189
|
Fcgrt
|
Fc receptor, IgG, alpha chain transporter
|
|
chr17_-_84246141
|
12.877
|
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr16_-_23029138
|
12.855
|
|
Kng2
|
kininogen 2
|
|
chr9_+_46049140
|
12.813
|
|
Apoa4
|
apolipoprotein A-IV
|
|
chr8_+_11313002
|
12.812
|
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr10_-_79446784
|
12.791
|
|
ORF61
|
open reading frame 61
|
|
chr15_-_89011113
|
12.719
|
NM_138749
|
Plxnb2
|
plexin B2
|