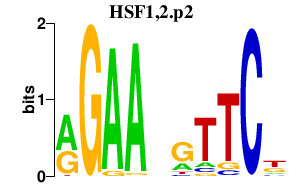
|
chr5_+_136363914
|
72.362
|
|
Hspb1
|
heat shock protein 1
|
|
chr11_-_55208276
|
50.467
|
|
|
|
|
chrX_+_54032914
|
43.227
|
|
Fhl1
|
four and a half LIM domains 1
|
|
chr1_-_172018982
|
41.571
|
NM_022563
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chrX_+_54032858
|
40.579
|
NM_001077362
|
Fhl1
|
four and a half LIM domains 1
|
|
chr1_-_66981900
|
40.150
|
NM_001113387
|
Myl1
|
myosin, light polypeptide 1
|
|
chr5_+_136363810
|
38.667
|
|
Hspb1
|
heat shock protein 1
|
|
chr14_-_46149660
|
38.317
|
NM_146054
|
Fermt2
|
fermitin family homolog 2 (Drosophila)
|
|
chr7_-_74879894
|
35.907
|
|
Synm
|
synemin, intermediate filament protein
|
|
chr4_+_41774206
|
34.150
|
NM_011335
NM_001193668
NM_001193666
NM_023052
|
Ccl21b
Gm10591
Gm13304
Ccl21a
Ccl21c
|
chemokine (C-C motif) ligand 21B (leucine)
predicted gene 10591
predicted gene 13304
chemokine (C-C motif) ligand 21A (serine)
chemokine (C-C motif) ligand 21C (leucine)
|
|
chr5_+_136363769
|
33.460
|
NM_013560
|
Hspb1
|
heat shock protein 1
|
|
chr1_+_166726827
|
31.212
|
NM_019759
|
Dpt
|
dermatopontin
|
|
chr7_+_147949708
|
31.165
|
NM_021282
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr7_+_147949637
|
30.387
|
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr4_-_82151004
|
29.701
|
|
Nfib
|
nuclear factor I/B
|
|
chr1_-_45559904
|
28.665
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr15_-_76881097
|
27.973
|
NM_013593
|
Mb
|
myoglobin
|
|
chr5_+_17080553
|
27.678
|
NM_013657
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr1_-_45559866
|
27.554
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr5_-_116872852
|
27.256
|
NM_030704
|
Hspb8
|
heat shock protein 8
|
|
chr4_-_42393043
|
27.097
|
NM_011335
NM_001193668
NM_001193666
NM_023052
|
Ccl21b
Gm10591
Gm13304
Ccl21a
Ccl21c
|
chemokine (C-C motif) ligand 21B (leucine)
predicted gene 10591
predicted gene 13304
chemokine (C-C motif) ligand 21A (serine)
chemokine (C-C motif) ligand 21C (leucine)
|
|
chr4_+_42231042
|
26.984
|
NM_001193667
NM_011124
|
Gm1987
Ccl21a
|
predicted gene 1987
chemokine (C-C motif) ligand 21A (serine)
|
|
chr4_-_42787449
|
26.961
|
NM_001193667
NM_011124
|
Gm1987
Ccl21a
|
predicted gene 1987
chemokine (C-C motif) ligand 21A (serine)
|
|
chr4_-_82151590
|
25.233
|
|
Nfib
|
nuclear factor I/B
|
|
chr16_+_45093784
|
25.186
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr7_-_31861665
|
24.683
|
NM_008557
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3
|
|
chr3_+_67696141
|
23.971
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr14_+_102239673
|
22.702
|
|
Lmo7
|
LIM domain only 7
|
|
chr2_-_125142715
|
22.377
|
|
Fbn1
|
fibrillin 1
|
|
chr7_-_106501642
|
21.589
|
NM_001111043
NM_001111044
NM_009825
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr7_-_106501620
|
21.224
|
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr1_-_82232392
|
21.056
|
|
Irs1
|
insulin receptor substrate 1
|
|
chr9_-_66881744
|
20.822
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr3_-_10208575
|
20.553
|
NM_024406
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_-_106501584
|
20.284
|
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr8_+_95376459
|
19.999
|
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr9_+_50561092
|
19.678
|
|
Cryab
|
crystallin, alpha B
|
|
chr11_+_66984528
|
19.275
|
NM_001039545
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult
|
|
chr6_+_17290478
|
18.884
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr14_+_20570444
|
18.637
|
NM_008695
|
Nid2
|
nidogen 2
|
|
chr17_-_35096167
|
18.488
|
NM_010478
|
Hspa1b
Hspa1a
|
heat shock protein 1B
heat shock protein 1A
|
|
chr1_-_79855177
|
18.391
|
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr3_-_126646303
|
18.174
|
NM_001034168
|
Ank2
|
ankyrin 2, brain
|
|
chr5_+_92760866
|
17.535
|
NM_181728
|
Art3
|
ADP-ribosyltransferase 3
|
|
chr12_+_76409299
|
17.377
|
NM_023275
|
Rhoj
|
ras homolog gene family, member J
|
|
chr4_-_82151657
|
17.339
|
|
Nfib
|
nuclear factor I/B
|
|
chr10_-_27336648
|
17.270
|
NM_008481
|
Lama2
|
laminin, alpha 2
|
|
chr1_+_43787442
|
16.890
|
NM_024283
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr16_-_45844409
|
16.738
|
NM_153412
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr18_-_43552694
|
16.283
|
NM_009468
|
Dpysl3
|
dihydropyrimidinase-like 3
|
|
chr14_+_51710846
|
16.224
|
|
Rnase4
|
ribonuclease, RNase A family 4
|
|
chr18_-_35881144
|
16.085
|
NM_001033141
|
Ecscr
|
endothelial cell-specific chemotaxis regulator
|
|
chr7_-_149762551
|
16.035
|
|
H19
|
H19 fetal liver mRNA
|
|
chr9_+_50561121
|
15.400
|
|
Cryab
|
crystallin, alpha B
|
|
chr1_+_43787537
|
15.385
|
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr12_+_110782654
|
15.263
|
|
Meg3
|
maternally expressed 3
|
|
chr9_+_50561315
|
15.245
|
|
Cryab
|
crystallin, alpha B
|
|
chr7_+_135667279
|
14.805
|
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr15_+_78673024
|
14.772
|
NM_027219
|
Cdc42ep1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr3_+_137004041
|
14.700
|
NM_001163522
NM_016885
|
Emcn
|
endomucin
|
|
chr1_+_43787692
|
14.662
|
|
1500015O10Rik
|
RIKEN cDNA 1500015O10 gene
|
|
chr9_+_54547344
|
14.529
|
NM_021422
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr6_+_4471231
|
14.467
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr9_-_54407955
|
14.415
|
|
Cib2
|
calcium and integrin binding family member 2
|
|
chr11_-_11832366
|
14.415
|
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr16_+_48842663
|
14.390
|
NM_020509
|
Retnla
|
resistin like alpha
|
|
chr6_-_136824217
|
14.279
|
|
Mgp
|
matrix Gla protein
|
|
chr18_+_20823750
|
14.176
|
NM_013697
|
Ttr
|
transthyretin
|
|
chr6_-_21802514
|
14.122
|
NM_173007
|
Tspan12
|
tetraspanin 12
|
|
chr12_+_110783593
|
14.107
|
|
Meg3
|
maternally expressed 3
|
|
chr11_+_53915017
|
14.001
|
|
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide
|
|
chr3_+_137004089
|
13.838
|
|
Emcn
|
endomucin
|
|
chr16_+_45093816
|
13.580
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr7_-_133606617
|
13.487
|
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr5_+_32913539
|
13.438
|
NM_009535
|
Yes1
|
Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
|
|
chr16_-_45844343
|
13.381
|
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr2_+_157385835
|
13.359
|
NM_010923
NM_180960
|
Nnat
|
neuronatin
|
|
chr1_+_137648101
|
13.353
|
|
Csrp1
|
cysteine and glycine-rich protein 1
|
|
chr6_+_34548512
|
13.241
|
|
Cald1
|
caldesmon 1
|
|
chr9_+_50561381
|
13.155
|
|
Cryab
|
crystallin, alpha B
|
|
chr6_+_56828743
|
13.102
|
|
Fkbp9
|
FK506 binding protein 9
|
|
chr7_-_112630662
|
12.980
|
NM_028444
|
Prkcdbp
|
protein kinase C, delta binding protein
|
|
chr5_+_93032108
|
12.916
|
|
Stbd1
|
starch binding domain 1
|
|
chr18_+_20823908
|
12.912
|
|
Ttr
|
transthyretin
|
|
chr9_+_54547190
|
12.869
|
|
Dnaja4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr7_+_119822831
|
12.773
|
NM_001166584
NM_009346
|
Tead1
|
TEA domain family member 1
|
|
chr7_+_135390325
|
12.622
|
NM_009365
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr1_-_45560001
|
12.550
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_-_79855209
|
12.502
|
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr10_+_18246228
|
12.392
|
|
Nhsl1
|
NHS-like 1
|
|
chr15_+_97962651
|
12.384
|
|
|
|
|
chr11_-_69761489
|
12.372
|
NM_009204
|
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr5_+_93032061
|
12.326
|
NM_175096
|
Stbd1
|
starch binding domain 1
|
|
chr2_+_20431673
|
12.190
|
NM_029895
NM_178059
|
Etl4
|
enhancer trap locus 4
|
|
chr2_+_122607734
|
12.095
|
NM_021507
|
Sqrdl
|
sulfide quinone reductase-like (yeast)
|
|
chr7_-_133606586
|
11.785
|
NM_007504
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chrX_-_73088288
|
11.714
|
NM_001166454
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr4_+_42625654
|
11.680
|
NM_011335
NM_001193668
NM_001193666
NM_023052
|
Ccl21b
Gm10591
Gm13304
Ccl21a
Ccl21c
|
chemokine (C-C motif) ligand 21B (leucine)
predicted gene 10591
predicted gene 13304
chemokine (C-C motif) ligand 21A (serine)
chemokine (C-C motif) ligand 21C (leucine)
|
|
chr4_-_82150687
|
11.675
|
|
Nfib
|
nuclear factor I/B
|
|
chr6_-_142456402
|
11.573
|
NM_008492
|
Ldhb
|
lactate dehydrogenase B
|
|
chr19_+_20676453
|
11.451
|
NM_013467
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1
|
|
chr7_+_119902897
|
11.434
|
NM_001166585
|
Tead1
|
TEA domain family member 1
|
|
chr17_-_56419161
|
11.356
|
|
Plin3
|
perilipin 3
|
|
chr10_+_79174315
|
11.334
|
|
Bsg
|
basigin
|
|
chr13_-_93564652
|
11.262
|
NM_011582
|
Thbs4
|
thrombospondin 4
|
|
chr19_+_38598686
|
11.243
|
NM_019588
|
Plce1
|
phospholipase C, epsilon 1
|
|
chr18_+_37896589
|
11.120
|
NM_033592
|
Pcdhga9
|
protocadherin gamma subfamily A, 9
|
|
chr17_+_44325518
|
11.087
|
NM_172621
|
Clic5
|
chloride intracellular channel 5
|
|
chr3_+_95965959
|
11.076
|
|
Mtmr11
|
myotubularin related protein 11
|
|
chr1_-_72921438
|
11.050
|
NM_010518
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr1_+_159436875
|
10.990
|
NM_033354
|
Sec16b
|
SEC16 homolog B (S. cerevisiae)
|
|
chr10_+_79353650
|
10.961
|
|
Cfd
|
complement factor D (adipsin)
|
|
chr6_+_30589440
|
10.918
|
|
Cpa1
|
carboxypeptidase A1
|
|
chr5_-_44485951
|
10.750
|
NM_001163578
NM_001163581
NM_001163582
NM_001163583
NM_001163584
NM_001163585
|
Prom1
|
prominin 1
|
|
chr2_+_35111861
|
10.737
|
|
Gsn
|
gelsolin
|
|
chr2_-_84614960
|
10.696
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr6_-_142519807
|
10.685
|
NM_008428
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8
|
|
chr9_+_50560862
|
10.672
|
NM_009964
|
Cryab
|
crystallin, alpha B
|
|
chr18_+_32091098
|
10.645
|
NM_144862
|
Lims2
|
LIM and senescent cell antigen like domains 2
|
|
chr16_+_30065419
|
10.624
|
NM_008235
|
Hes1
|
hairy and enhancer of split 1 (Drosophila)
|
|
chr10_+_79174337
|
10.558
|
|
Bsg
|
basigin
|
|
chrX_+_10173557
|
10.509
|
|
|
|
|
chr11_+_78002569
|
10.475
|
|
Rab34
|
RAB34, member of RAS oncogene family
|
|
chr4_+_150229178
|
10.456
|
NM_133753
|
Errfi1
|
ERBB receptor feedback inhibitor 1
|
|
chr14_+_51710751
|
10.437
|
NM_001161731
NM_021472
|
Ang
Rnase4
|
angiogenin, ribonuclease, RNase A family, 5
ribonuclease, RNase A family 4
|
|
chr1_+_94728419
|
10.365
|
|
Gpc1
|
glypican 1
|
|
chr5_-_150438704
|
10.290
|
|
Hsph1
|
heat shock 105kDa/110kDa protein 1
|
|
chr1_-_72921098
|
10.239
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr7_-_86887849
|
10.184
|
NM_011068
|
Pex11a
|
peroxisomal biogenesis factor 11 alpha
|
|
chr4_-_58083082
|
10.076
|
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr2_+_152561983
|
10.072
|
NM_010495
|
Id1
|
inhibitor of DNA binding 1
|
|
chr5_-_150438750
|
10.070
|
|
Hsph1
|
heat shock 105kDa/110kDa protein 1
|
|
chr18_-_14002490
|
10.068
|
|
Zfp521
|
zinc finger protein 521
|
|
chr6_-_21802087
|
9.938
|
|
Tspan12
|
tetraspanin 12
|
|
chr18_+_23573939
|
9.929
|
|
Dtna
|
dystrobrevin alpha
|
|
chr7_-_109082300
|
9.812
|
NM_001162973
NM_027053
|
Lrrc51
|
leucine rich repeat containing 51
|
|
chr7_-_4704436
|
9.777
|
NM_183406
NM_183405
|
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2
|
|
chr2_-_84614976
|
9.697
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr11_+_28767772
|
9.612
|
|
Efemp1
|
epidermal growth factor-containing fibulin-like extracellular matrix protein 1
|
|
chr7_+_134355216
|
9.526
|
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr4_+_133871031
|
9.439
|
NM_001039048
|
Trim63
|
tripartite motif-containing 63
|
|
chr15_-_96886327
|
9.390
|
NM_027052
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr1_-_72920776
|
9.367
|
|
Igfbp5
|
insulin-like growth factor binding protein 5
|
|
chr8_-_47292973
|
9.358
|
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4
|
|
chr3_+_95965892
|
9.351
|
NM_181409
|
Mtmr11
|
myotubularin related protein 11
|
|
chr11_+_98245124
|
9.320
|
NM_011540
|
Tcap
|
titin-cap
|
|
chr11_+_96711152
|
9.294
|
NM_019877
|
Copz2
|
coatomer protein complex, subunit zeta 2
|
|
chr9_-_54407862
|
9.245
|
NM_019686
|
Cib2
|
calcium and integrin binding family member 2
|
|
chrX_+_93650774
|
9.161
|
NM_001159628
NM_010417
|
Heph
|
hephaestin
|
|
chr7_+_135689619
|
9.134
|
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr9_-_101101334
|
9.056
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr9_-_15105965
|
9.041
|
NM_133732
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr12_+_32008372
|
9.039
|
|
Lamb1
|
laminin B1
|
|
chr10_-_75106702
|
8.977
|
NM_001162913
NM_027890
|
Susd2
|
sushi domain containing 2
|
|
chr2_+_172978566
|
8.851
|
NM_011044
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic
|
|
chr17_+_44325664
|
8.847
|
|
Clic5
|
chloride intracellular channel 5
|
|
chr14_+_69984344
|
8.831
|
|
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4
|
|
chr4_-_43536296
|
8.824
|
|
Tpm2
|
tropomyosin 2, beta
|
|
chr2_+_164388211
|
8.790
|
NM_026323
|
Wfdc2
|
WAP four-disulfide core domain 2
|
|
chrX_+_132528317
|
8.775
|
NM_029823
|
6530401D17Rik
2900062L11Rik
|
RIKEN cDNA 6530401D17 gene
RIKEN cDNA 2900062L11 gene
|
|
chr10_-_78054639
|
8.698
|
NM_027875
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans)
|
|
chr3_+_90346956
|
8.674
|
|
S100a16
|
S100 calcium binding protein A16
|
|
chr1_+_34347244
|
8.544
|
|
Dst
|
dystonin
|
|
chr6_+_81991613
|
8.538
|
NM_145570
|
Fam176a
|
family with sequence similarity 176, member A
|
|
chr18_+_23573908
|
8.496
|
NM_010087
NM_207650
|
Dtna
|
dystrobrevin alpha
|
|
chr19_-_20802015
|
8.451
|
NM_011921
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7
|
|
chr15_+_100165359
|
8.433
|
NM_001081471
|
Gm10035
|
predicted gene 10035
|
|
chr14_-_122196929
|
8.388
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr16_+_48872720
|
8.382
|
NM_181596
|
Retnlg
|
resistin like gamma
|
|
chr18_+_23573977
|
8.377
|
|
Dtna
|
dystrobrevin alpha
|
|
chr1_-_174221511
|
8.329
|
|
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr4_+_135700717
|
8.308
|
|
Id3
|
inhibitor of DNA binding 3
|
|
chr7_-_77511607
|
8.279
|
NM_183261
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr9_-_30957394
|
8.277
|
|
|
|
|
chr17_+_34807445
|
8.240
|
NM_031176
|
Tnxb
|
tenascin XB
|
|
chr7_+_99710002
|
8.210
|
|
Ccdc90b
|
coiled-coil domain containing 90B
|
|
chr8_+_47576483
|
8.196
|
|
Acsl1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr4_-_82151180
|
8.096
|
NM_001113209
NM_001113210
NM_008687
|
Nfib
|
nuclear factor I/B
|
|
chr7_+_135667095
|
8.024
|
NM_013863
|
Bag3
|
BCL2-associated athanogene 3
|
|
chr18_-_53577625
|
7.997
|
NM_008908
|
Ppic
|
peptidylprolyl isomerase C
|
|
chrM_+_12383
|
7.965
|
|
|
|
|
chr9_+_14080805
|
7.916
|
|
Sesn3
|
sestrin 3
|
|
chr9_+_14080744
|
7.888
|
NM_030261
|
Sesn3
|
sestrin 3
|
|
chr2_+_174123359
|
7.874
|
NM_001077507
NM_010309
NM_201617
NM_201618
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus
|
|
chr9_-_15105864
|
7.843
|
|
4931406C07Rik
|
RIKEN cDNA 4931406C07 gene
|
|
chr11_+_100277007
|
7.826
|
NM_001163481
NM_010221
|
Fkbp10
|
FK506 binding protein 10
|
|
chr19_+_58744854
|
7.823
|
NM_026925
|
Pnlip
|
pancreatic lipase
|
|
chr10_-_126507271
|
7.802
|
NM_025982
|
Tspan31
|
tetraspanin 31
|
|
chr2_-_58419236
|
7.738
|
NM_001110204
NM_001110205
NM_007394
|
Acvr1
|
activin A receptor, type 1
|
|
chr5_+_93112640
|
7.701
|
|
Shroom3
|
shroom family member 3
|
|
chr7_-_4467556
|
7.681
|
NM_011618
|
Tnnt1
|
troponin T1, skeletal, slow
|
|
chr7_+_99709652
|
7.654
|
NM_001162918
NM_025515
|
Ccdc90b
|
coiled-coil domain containing 90B
|
|
chr5_+_97222355
|
7.612
|
NM_013470
|
Anxa3
|
annexin A3
|
|
chr8_-_47292863
|
7.478
|
|
|
|
|
chr7_+_146574969
|
7.443
|
NM_001127363
NM_183144
|
Inpp5a
|
inositol polyphosphate-5-phosphatase A
|
|
chr6_+_138089041
|
7.437
|
NM_019946
|
Mgst1
|
microsomal glutathione S-transferase 1
|
|
chrX_+_70728994
|
7.385
|
|
Bgn
|
biglycan
|
|
chr13_+_49639799
|
7.363
|
NM_001172481
NM_025711
|
Aspn
|
asporin
|
|
chr3_-_10208480
|
7.328
|
|
Fabp4
Fabp9
|
fatty acid binding protein 4, adipocyte
fatty acid binding protein 9, testis
|
|
chr1_+_173155184
|
7.310
|
NM_013474
|
Apoa2
|
apolipoprotein A-II
|