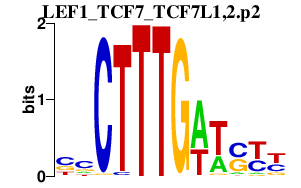
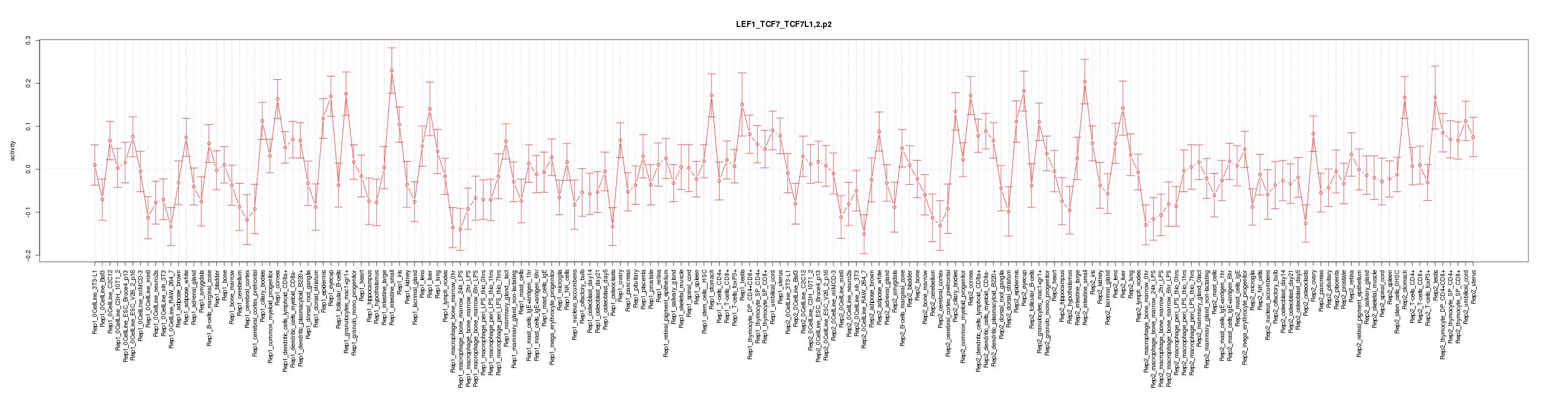
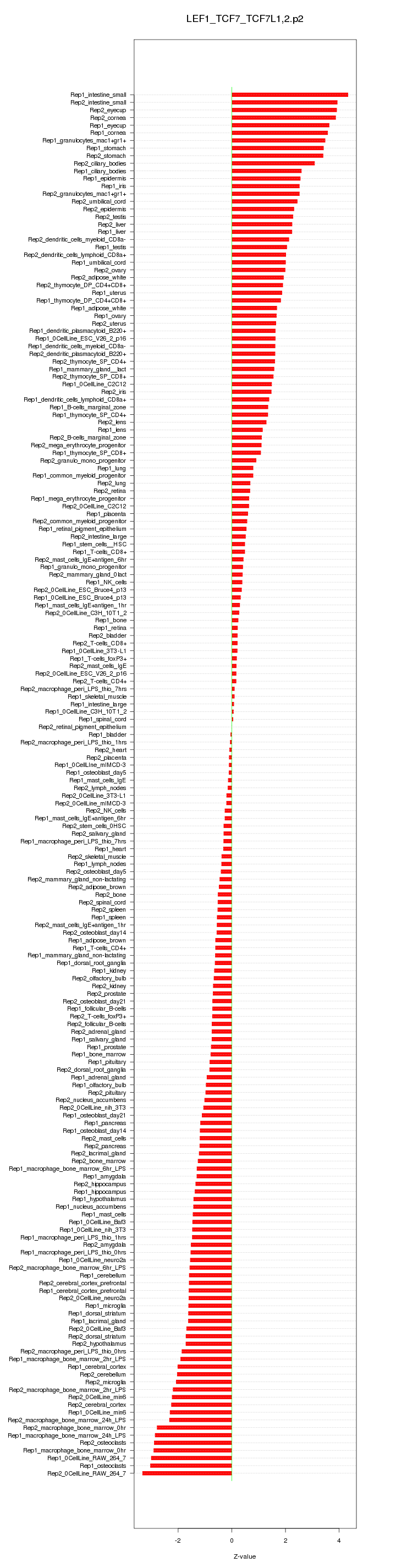
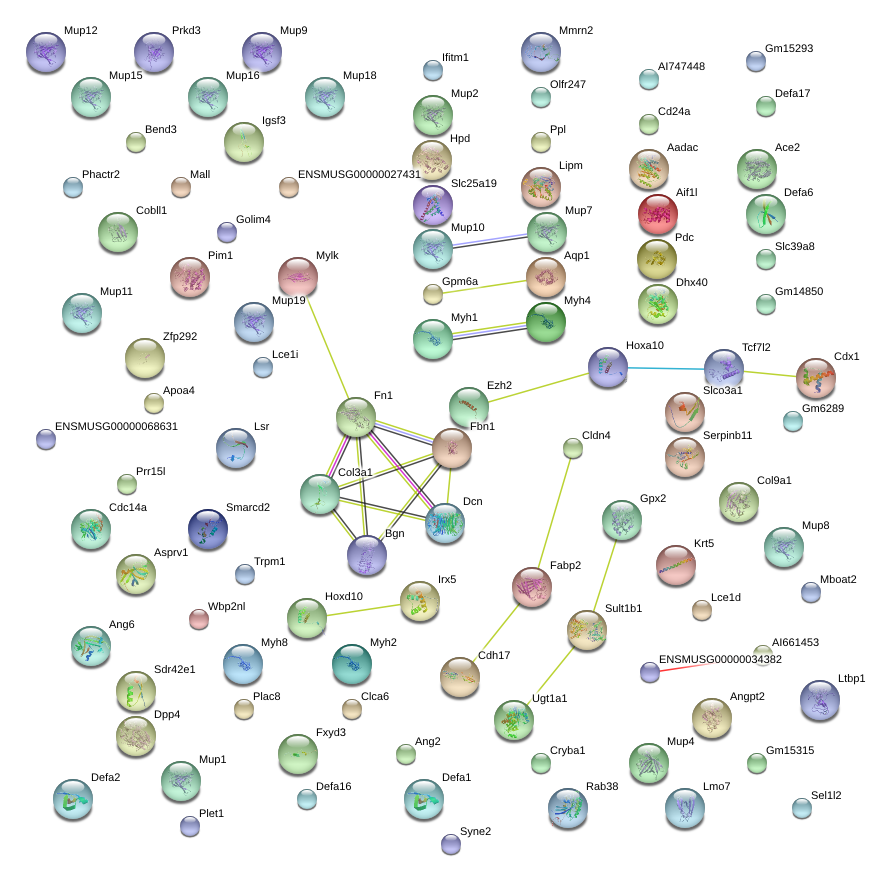
|
chr6_+_86578225
|
17.806
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr6_+_86578167
|
17.273
|
NM_026414
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr6_+_86578269
|
14.454
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr6_+_86578550
|
13.893
|
|
Asprv1
|
aspartic peptidase, retroviral-like 1
|
|
chr11_-_77538789
|
12.471
|
NM_009965
|
Cryba1
|
crystallin, beta A1
|
|
chr14_-_44579933
|
12.440
|
NM_001011876
|
Ang6
|
angiogenin, ribonuclease A family, member 6
|
|
chr10_+_96944961
|
11.349
|
NM_007833
|
Dcn
|
decorin
|
|
chr16_+_35002066
|
10.667
|
|
Mylk
|
myosin, light polypeptide kinase
|
|
chr1_+_45401303
|
10.300
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chrX_+_70728994
|
10.093
|
|
Bgn
|
biglycan
|
|
chrX_+_70728939
|
10.028
|
NM_007542
|
Bgn
|
biglycan
|
|
chr2_-_125142715
|
9.998
|
|
Fbn1
|
fibrillin 1
|
|
chr10_+_43198945
|
9.597
|
NM_199028
|
Bend3
|
BEN domain containing 3
|
|
chr10_+_96945057
|
9.505
|
|
Dcn
|
decorin
|
|
chr6_+_55286292
|
9.356
|
NM_007472
|
Aqp1
|
aquaporin 1
|
|
chr3_-_92491103
|
9.200
|
NM_027137
|
Lce1d
|
late cornified envelope 1D
|
|
chr3_+_101266000
|
9.164
|
|
Igsf3
|
immunoglobulin superfamily, member 3
|
|
chr5_-_135422730
|
8.868
|
|
Cldn4
|
claudin 4
|
|
chr7_-_31861665
|
8.846
|
NM_008557
|
Fxyd3
|
FXYD domain-containing ion transport regulator 3
|
|
chr5_-_135422795
|
8.749
|
NM_009903
|
Cldn4
|
claudin 4
|
|
chr3_+_122598292
|
8.546
|
NM_007980
|
Fabp2
|
fatty acid binding protein 2, intestinal
|
|
chr1_+_109258889
|
8.317
|
NM_025867
|
Serpinb11
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 11
|
|
chr5_-_135422780
|
8.287
|
|
Cldn4
|
claudin 4
|
|
chr4_-_34830178
|
8.238
|
NM_013889
|
Zfp292
|
zinc finger protein 292
|
|
chr1_+_59543450
|
8.170
|
|
|
|
|
chr7_+_95578782
|
8.068
|
NM_028238
|
Rab38
|
RAB38, member of RAS oncogene family
|
|
chr8_+_22675857
|
8.041
|
NM_001177522
|
Gm14850
|
predicted gene 14850
|
|
chr1_+_45401659
|
8.022
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr15_-_58010344
|
7.953
|
|
|
|
|
chr7_+_71330344
|
7.780
|
NM_018752
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1
|
|
chr9_+_50302624
|
7.720
|
NM_029639
|
1600029D21Rik
|
RIKEN cDNA 1600029D21 gene
|
|
chr16_-_5132502
|
7.705
|
NM_008909
|
Ppl
|
periplakin
|
|
chr2_+_31805852
|
7.669
|
|
Aif1l
|
allograft inflammatory factor 1-like
|
|
chr1_+_152166546
|
7.652
|
NM_001159730
NM_024458
|
Pdc
|
phosducin
|
|
chr2_-_127555628
|
7.615
|
NM_145532
|
Mall
|
mal, T-cell differentiation protein-like
|
|
chr17_+_75404893
|
7.486
|
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr4_-_60594469
|
7.382
|
|
Mup1
|
major urinary protein 1
|
|
chr17_+_29629987
|
7.282
|
|
Pim1
|
proviral integration site 1
|
|
chr6_-_47544925
|
7.208
|
NM_001146689
NM_007971
|
Ezh2
|
enhancer of zeste homolog 2 (Drosophila)
|
|
chr17_+_47605313
|
7.199
|
|
AI661453
|
expressed sequence AI661453
|
|
chr12_+_25516433
|
7.198
|
NM_001083341
NM_026037
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr14_+_35188650
|
7.041
|
NM_153127
|
Mmrn2
|
multimerin 2
|
|
chr10_+_43298969
|
6.988
|
NM_009846
|
Cd24a
|
CD24a antigen
|
|
chr8_+_22766238
|
6.862
|
NM_001167790
|
Defa3
Defa17
|
defensin, alpha, 3
defensin, alpha, 17
|
|
chr18_-_61195852
|
6.839
|
NM_009880
|
Cdx1
|
caudal type homeobox 1
|
|
chr7_-_31758185
|
6.776
|
|
Lsr
|
lipolysis stimulated lipoprotein receptor
|
|
chr2_-_65076471
|
6.753
|
|
Cobll1
|
Cobl-like 1
|
|
chr14_+_102239673
|
6.699
|
|
Lmo7
|
LIM domain only 7
|
|
chr14_+_102129129
|
6.695
|
NM_201529
|
Lmo7
|
LIM domain only 7
|
|
chr17_+_75404865
|
6.630
|
NM_019919
|
Ltbp1
|
latent transforming growth factor beta binding protein 1
|
|
chr3_-_92582820
|
6.585
|
NM_029667
|
Lce1i
|
late cornified envelope 1I
|
|
chr5_-_101001209
|
6.580
|
NM_139198
|
Plac8
|
placenta-specific 8
|
|
chr10_-_13108775
|
6.570
|
NM_001195066
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr5_-_123632567
|
6.563
|
NM_008277
|
Hpd
|
4-hydroxyphenylpyruvic acid dioxygenase
|
|
chr8_+_22844965
|
6.524
|
NM_007852
NM_001024225
|
Defa3
Defa6
Defa24
|
defensin, alpha, 3
defensin, alpha, 6
defensin, alpha, 24
|
|
chr5_-_101001191
|
6.518
|
|
Plac8
|
placenta-specific 8
|
|
chr2_-_65076584
|
6.466
|
NM_027225
NM_177025
|
Cobll1
|
Cobl-like 1
|
|
chr4_+_11685294
|
6.303
|
NM_019753
|
Cdh17
|
cadherin 17
|
|
chr3_-_116126932
|
6.294
|
NM_001080818
NM_001173553
|
Cdc14a
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chr4_-_60594496
|
6.264
|
|
Mup1
|
major urinary protein 1
|
|
chr3_-_144595486
|
6.226
|
NM_001033199
|
AI747448
|
expressed sequence AI747448
|
|
chr11_-_116668593
|
6.222
|
|
|
|
|
chr12_-_77896363
|
6.189
|
|
Gpx2
|
glutathione peroxidase 2
|
|
chr7_-_81429262
|
6.184
|
|
Slco3a1
|
solute carrier organic anion transporter family, member 3a1
|
|
chr12_+_77182878
|
6.085
|
|
Syne2
|
synaptic nuclear envelope 2
|
|
chr9_+_46048812
|
6.084
|
NM_007468
|
Apoa4
|
apolipoprotein A-IV
|
|
chr19_+_55969810
|
6.069
|
NM_001142924
|
Tcf7l2
|
transcription factor 7-like 2, T-cell specific, HMG-box
|
|
chr1_-_71686889
|
6.037
|
|
Fn1
|
fibronectin 1
|
|
chr3_-_144637974
|
5.950
|
NM_207208
|
Clca6
|
chloride channel calcium activated 6
|
|
chr5_-_87964170
|
5.948
|
|
Sult1b1
|
sulfotransferase family 1B, member 1
|
|
chr9_+_46048993
|
5.851
|
|
Apoa4
|
apolipoprotein A-IV
|
|
chr6_-_52190771
|
5.841
|
NM_001122950
|
Hoxa10
|
homeobox A10
|
|
chr2_-_62249869
|
5.828
|
|
Dpp4
|
dipeptidylpeptidase 4
|
|
chr3_+_59835700
|
5.797
|
NM_023383
|
Aadac
|
arylacetamide deacetylase (esterase)
|
|
chr7_+_148153974
|
5.776
|
NM_001112715
|
Ifitm1
|
interferon induced transmembrane protein 1
|
|
chrX_+_160600108
|
5.766
|
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr12_-_77896410
|
5.662
|
|
Gpx2
|
glutathione peroxidase 2
|
|
chr12_-_77896534
|
5.655
|
NM_030677
|
Gpx2
|
glutathione peroxidase 2
|
|
chr12_+_31950252
|
5.621
|
|
Lamb1
|
laminin B1
|
|
chr7_+_148153327
|
5.612
|
NM_026820
|
Ifitm1
|
interferon induced transmembrane protein 1
|
|
chr19_+_34175422
|
5.590
|
NM_023903
|
Lipm
|
lipase, family member M
|
|
chr11_-_106128935
|
5.576
|
NM_031878
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2
|
|
chr17_-_79420125
|
5.477
|
NM_001171004
NM_029239
|
Prkd3
|
protein kinase D3
|
|
chr2_+_144649401
|
5.466
|
NM_025490
|
1700010M22Rik
|
RIKEN cDNA 1700010M22 gene
|
|
chr15_-_101543318
|
5.436
|
NM_027011
|
Krt5
|
keratin 5
|
|
chr1_+_90062617
|
5.424
|
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr14_-_51815597
|
5.371
|
NM_007449
|
Ang2
|
angiogenin, ribonuclease A family, member 2
|
|
chr1_+_90062550
|
5.318
|
NM_201643
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5
|
|
chr3_-_75760820
|
5.286
|
|
Golim4
|
golgi integral membrane protein 4
|
|
chr11_+_96790581
|
5.286
|
NM_146026
|
Prr15l
|
proline rich 15-like
|
|
chr2_-_140215417
|
5.247
|
NM_001033296
|
Sel1l2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr11_-_86621130
|
5.240
|
NM_026191
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr3_+_135488617
|
5.232
|
NM_001135149
NM_001135150
|
Slc39a8
|
solute carrier family 39 (metal ion transporter), member 8
|
|
chr1_+_24184448
|
5.214
|
NM_007740
|
Col9a1
|
collagen, type IX, alpha 1
|
|
chr4_+_11685359
|
5.211
|
|
Cdh17
|
cadherin 17
|
|
chr11_-_86621074
|
5.156
|
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40
|
|
chr8_-_120195414
|
5.131
|
NM_028725
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1
|
|
chr15_+_82129383
|
5.128
|
NM_029066
|
Wbp2nl
|
WBP2 N-terminal like
|
|
chr8_+_94881694
|
5.077
|
NM_018826
|
Irx5
|
Iroquois related homeobox 5 (Drosophila)
|
|
chr11_+_67013553
|
5.048
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr3_-_57379600
|
4.991
|
NM_133784
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr4_-_61179982
|
4.982
|
|
Mup1
|
major urinary protein 1
|
|
chr12_+_31950097
|
4.963
|
NM_008482
|
Lamb1
|
laminin B1
|
|
chr3_-_92474213
|
4.942
|
NM_028625
|
Lce1a2
|
late cornified envelope 1A2
|
|
chr3_+_96025040
|
4.911
|
NM_178214
|
Hist2h2be
|
histone cluster 2, H2be
|
|
chr3_-_40659202
|
4.911
|
NM_001129886
|
Gm10731
|
predicted gene 10731
|
|
chr6_+_68375560
|
4.828
|
|
Igkv16-104
|
immunoglobulin kappa variable 16-104
|
|
chr5_-_105539282
|
4.827
|
NM_172777
|
Gbp9
|
guanylate-binding protein 9
|
|
chr10_+_79451332
|
4.819
|
NM_007725
|
Cnn2
|
calponin 2
|
|
chr17_+_28667805
|
4.769
|
NM_026290
|
4930511I11Rik
|
RIKEN cDNA 4930511I11 gene
|
|
chr1_-_58752798
|
4.741
|
NM_175370
|
Als2cr12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 (human)
|
|
chr10_-_126807486
|
4.727
|
NM_010565
|
Inhbc
|
inhibin beta-C
|
|
chr10_-_61441817
|
4.694
|
|
Col13a1
|
collagen, type XIII, alpha 1
|
|
chr17_-_84246141
|
4.689
|
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr6_+_29685467
|
4.654
|
NM_176996
|
Smo
|
smoothened homolog (Drosophila)
|
|
chr17_-_84246073
|
4.652
|
NM_025325
|
Haao
|
3-hydroxyanthranilate 3,4-dioxygenase
|
|
chr13_-_3803547
|
4.649
|
NM_027416
|
Calml3
|
calmodulin-like 3
|
|
chr11_+_102006824
|
4.640
|
|
Nags
|
N-acetylglutamate synthase
|
|
chr7_-_111161213
|
4.630
|
NM_147084
|
Olfr639
|
olfactory receptor 639
|
|
chr15_-_58046380
|
4.598
|
|
Fbxo32
|
F-box protein 32
|
|
chr4_-_58219368
|
4.558
|
NM_022814
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1
|
|
chr13_-_41454331
|
4.526
|
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr10_-_61441842
|
4.522
|
NM_007731
|
Col13a1
|
collagen, type XIII, alpha 1
|
|
chr12_+_21117704
|
4.522
|
|
Asap2
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2
|
|
chr11_+_108781661
|
4.514
|
NM_015732
|
Axin2
|
axin2
|
|
chr11_+_102006847
|
4.453
|
|
Nags
|
N-acetylglutamate synthase
|
|
chr16_+_22951183
|
4.416
|
|
Hrg
|
histidine-rich glycoprotein
|
|
chr11_+_46218231
|
4.396
|
NM_001013783
|
Fam71b
|
family with sequence similarity 71, member B
|
|
chr1_+_36818669
|
4.386
|
NM_009539
|
Zap70
|
zeta-chain (TCR) associated protein kinase
|
|
chrX_-_130696466
|
4.385
|
NM_183319
|
Xkrx
|
X Kell blood group precursor related X linked
|
|
chr15_-_82624668
|
4.333
|
NM_029562
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26
|
|
chr14_-_98568744
|
4.305
|
NM_001038610
NM_007826
|
Dach1
|
dachshund 1 (Drosophila)
|
|
chr4_-_149377238
|
4.256
|
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase)
|
|
chr11_-_99949426
|
4.241
|
NM_001159374
|
Krt32
|
keratin 32
|
|
chr7_+_119167048
|
4.226
|
|
Usp47
|
ubiquitin specific peptidase 47
|
|
chr3_+_92736900
|
4.212
|
NM_025501
|
Lce3b
|
late cornified envelope 3B
|
|
chr7_+_147949637
|
4.202
|
|
Cyp2e1
|
cytochrome P450, family 2, subfamily e, polypeptide 1
|
|
chr3_-_57379831
|
4.194
|
NM_001168281
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr12_-_56087322
|
4.171
|
NM_013815
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A
|
|
chr5_+_32541456
|
4.165
|
NM_001081407
|
Plb1
|
phospholipase B1
|
|
chr1_+_193730645
|
4.161
|
NM_009579
|
Slc30a1
|
solute carrier family 30 (zinc transporter), member 1
|
|
chr11_+_87881747
|
4.159
|
NM_016686
|
Vezf1
|
vascular endothelial zinc finger 1
|
|
chr11_+_67051530
|
4.127
|
NM_010855
|
Myh4
|
myosin, heavy polypeptide 4, skeletal muscle
|
|
chr12_+_114378368
|
4.127
|
NM_024223
|
Crip2
|
cysteine rich protein 2
|
|
chr5_+_100408033
|
4.090
|
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene
|
|
chr19_+_20676453
|
4.071
|
NM_013467
|
Aldh1a1
|
aldehyde dehydrogenase family 1, subfamily A1
|
|
chr16_+_22951144
|
4.058
|
NM_053176
|
Hrg
|
histidine-rich glycoprotein
|
|
chr1_-_157589156
|
4.051
|
NM_010712
|
Lhx4
|
LIM homeobox protein 4
|
|
chr7_+_97535516
|
4.047
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr7_-_112936063
|
4.020
|
NM_001162943
|
Dchs1
|
dachsous 1 (Drosophila)
|
|
chr15_-_82624638
|
4.018
|
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26
|
|
chr13_+_49703402
|
4.016
|
NM_008760
|
Ogn
|
osteoglycin
|
|
chr14_+_27852148
|
4.013
|
NM_134437
|
Il17rd
|
interleukin 17 receptor D
|
|
chr7_-_123182257
|
4.009
|
NM_001025559
|
Sox6
|
SRY-box containing gene 6
|
|
chr3_-_92452189
|
3.977
|
NM_025984
|
Lce1a1
|
late cornified envelope 1A1
|
|
chr4_-_43496556
|
3.970
|
NM_019436
|
Sit1
|
suppression inducing transmembrane adaptor 1
|
|
chr16_+_45094163
|
3.965
|
NM_026439
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr14_+_49791899
|
3.933
|
NM_001081430
|
Naa30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit
|
|
chr14_+_22319075
|
3.924
|
NM_017479
|
Myst4
|
MYST histone acetyltransferase monocytic leukemia 4
|
|
chr3_+_92483168
|
3.914
|
NM_028622
|
Lce1c
|
late cornified envelope 1C
|
|
chr1_-_165243608
|
3.901
|
|
Prrx1
|
paired related homeobox 1
|
|
chr14_-_73762324
|
3.894
|
|
Itm2b
|
integral membrane protein 2B
|
|
chrX_+_160578411
|
3.885
|
NM_027286
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2
|
|
chr12_+_114378688
|
3.881
|
|
Crip2
|
cysteine rich protein 2
|
|
chr8_-_112486020
|
3.877
|
NM_028584
NM_212447
|
Marveld3
|
MARVEL (membrane-associating) domain containing 3
|
|
chr6_-_78418862
|
3.877
|
NM_011260
|
Reg3g
|
regenerating islet-derived 3 gamma
|
|
chr7_+_27904997
|
3.865
|
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr15_-_58046428
|
3.865
|
NM_026346
|
Fbxo32
|
F-box protein 32
|
|
chr12_+_18521315
|
3.858
|
NM_001033157
|
5730507C01Rik
|
RIKEN cDNA 5730507C01 gene
|
|
chr3_-_98567014
|
3.822
|
NM_001161742
NM_001161743
|
Hsd3b3
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 3
|
|
chr9_+_107897371
|
3.809
|
NM_001122635
|
Cdhr4
|
cadherin-related family member 4
|
|
chr2_-_143688908
|
3.777
|
NM_009751
|
Bfsp1
|
beaded filament structural protein 1, in lens-CP94
|
|
chr4_+_135842003
|
3.770
|
|
Zfp46
|
zinc finger protein 46
|
|
chr7_+_125743753
|
3.769
|
|
Tmc5
|
transmembrane channel-like gene family 5
|
|
chr3_-_116126853
|
3.769
|
|
Cdc14a
|
CDC14 cell division cycle 14 homolog A (S. cerevisiae)
|
|
chrX_-_35774388
|
3.752
|
|
Lamp2
|
lysosomal-associated membrane protein 2
|
|
chr7_+_97535490
|
3.719
|
NM_001040087
NM_001040088
|
Sytl2
|
synaptotagmin-like 2
|
|
chr14_-_118450875
|
3.710
|
NM_010024
|
Dct
|
dopachrome tautomerase
|
|
chr18_-_58169723
|
3.670
|
|
Fbn2
|
fibrillin 2
|
|
chr6_-_52184909
|
3.664
|
NM_008263
|
Hoxa10
|
homeobox A10
|
|
chr17_+_30763879
|
3.659
|
NM_013811
|
Dnahc8
|
dynein, axonemal, heavy chain 8
|
|
chr5_+_25021884
|
3.655
|
NM_198621
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1
|
|
chr4_-_132818988
|
3.643
|
NM_031393
|
Sytl1
|
synaptotagmin-like 1
|
|
chr15_-_75950624
|
3.640
|
NM_144848
|
Eppk1
BC024139
|
epiplakin 1
cDNA sequence BC024139
|
|
chr4_+_140976693
|
3.635
|
NM_013868
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr7_-_133819870
|
3.632
|
NM_133670
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1
|
|
chr12_-_104594210
|
3.629
|
NM_023049
|
Asb2
|
ankyrin repeat and SOCS box-containing 2
|
|
chr10_+_5959030
|
3.604
|
|
Zbtb2
|
zinc finger and BTB domain containing 2
|
|
chr1_+_92100572
|
3.601
|
NM_007722
|
Cxcr7
|
chemokine (C-X-C motif) receptor 7
|
|
chr15_-_98701613
|
3.590
|
NM_001033276
|
Mll2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr7_+_87343467
|
3.583
|
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B
|
|
chr4_+_43521765
|
3.553
|
|
Car9
|
carbonic anhydrase 9
|
|
chr19_-_39815590
|
3.545
|
NM_001039555
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68
|
|
chr2_+_174586283
|
3.541
|
|
Edn3
|
endothelin 3
|
|
chr16_+_45094196
|
3.513
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr4_+_43888373
|
3.511
|
NM_016678
|
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs
|
|
chr7_+_27904968
|
3.502
|
NM_007817
|
Cyp2f2
|
cytochrome P450, family 2, subfamily f, polypeptide 2
|
|
chr2_-_62249889
|
3.495
|
|
Dpp4
|
dipeptidylpeptidase 4
|
|
chr7_+_97497207
|
3.495
|
NM_001040085
NM_031394
|
Sytl2
|
synaptotagmin-like 2
|
|
chr16_+_35001358
|
3.486
|
|
Mylk
|
myosin, light polypeptide kinase
|