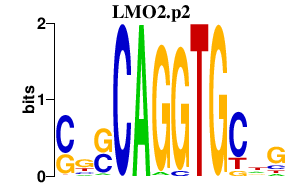
|
chr8_-_41720091
|
49.371
|
NM_001040699
|
Mtmr7
|
myotubularin related protein 7
|
|
chr8_+_128519001
|
43.646
|
NM_008430
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr8_+_128519295
|
40.662
|
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chrX_+_162676903
|
39.731
|
|
Gpm6b
|
glycoprotein m6b
|
|
chr14_-_55531726
|
39.465
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr15_+_78744862
|
35.528
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr4_-_133053867
|
32.869
|
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr6_+_118016385
|
32.865
|
NM_027526
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr11_-_69419330
|
31.989
|
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr14_-_30534966
|
29.891
|
NM_009785
|
Cacna2d3
|
calcium channel, voltage-dependent, alpha2/delta subunit 3
|
|
chr7_+_114514258
|
29.874
|
NM_021889
|
Syt9
|
synaptotagmin IX
|
|
chr2_+_49474840
|
29.324
|
|
Kif5c
|
kinesin family member 5C
|
|
chrX_+_162676870
|
29.002
|
NM_001177961
NM_001177962
NM_023122
|
Gpm6b
|
glycoprotein m6b
|
|
chr10_-_18463500
|
28.474
|
NM_001033258
|
D10Bwg1379e
|
DNA segment, Chr 10, Brigham & Women's Genetics 1379 expressed
|
|
chr12_+_110798661
|
27.050
|
|
Meg3
|
maternally expressed 3
|
|
chr2_+_157387284
|
26.808
|
|
Nnat
|
neuronatin
|
|
chr4_-_133054355
|
26.205
|
NM_001081156
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr6_-_23789048
|
25.390
|
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr7_+_52955153
|
24.985
|
NM_009800
|
Car11
|
carbonic anhydrase 11
|
|
chr18_+_82753560
|
24.204
|
|
Mbp
|
myelin basic protein
|
|
chr2_-_113669229
|
23.967
|
|
Scg5
|
secretogranin V
|
|
chr2_-_113669243
|
23.817
|
NM_009162
|
Scg5
|
secretogranin V
|
|
chr11_+_92960603
|
23.419
|
NM_028296
|
Car10
|
carbonic anhydrase 10
|
|
chr8_+_12915892
|
23.067
|
NM_178076
|
Mcf2l
|
mcf.2 transforming sequence-like
|
|
chr4_-_151235861
|
22.754
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr6_-_77929660
|
22.697
|
NM_001109764
NM_009819
NM_145732
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr2_-_113669159
|
22.381
|
|
Scg5
|
secretogranin V
|
|
chr15_+_78744802
|
22.348
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr2_-_127481868
|
22.051
|
|
Mal
|
myelin and lymphocyte protein, T-cell differentiation protein
|
|
chr7_+_52955569
|
21.433
|
|
Car11
|
carbonic anhydrase 11
|
|
chr5_-_44492908
|
21.375
|
NM_001163577
NM_008935
|
Prom1
|
prominin 1
|
|
chr5_-_68238603
|
21.319
|
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr2_+_49474775
|
21.304
|
NM_008449
|
Kif5c
|
kinesin family member 5C
|
|
chr6_-_77929543
|
21.281
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr6_-_25639812
|
21.241
|
|
Gpr37
|
G protein-coupled receptor 37
|
|
chr9_-_107134182
|
21.064
|
NM_153413
|
Dock3
|
dedicator of cyto-kinesis 3
|
|
chr15_+_78744576
|
20.790
|
|
Pdxp
|
pyridoxal (pyridoxine, vitamin B6) phosphatase
|
|
chr13_+_97841524
|
19.955
|
NM_001100458
|
Fam169a
|
family with sequence similarity 169, member A
|
|
chr6_-_25639835
|
19.809
|
|
Gpr37
|
G protein-coupled receptor 37
|
|
chr7_+_64845887
|
19.790
|
NM_001038701
|
Gabrb3
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 3
|
|
chr9_+_110147867
|
19.785
|
|
Cspg5
|
chondroitin sulfate proteoglycan 5
|
|
chr6_-_77929685
|
19.224
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr2_+_132607013
|
19.118
|
NM_007694
|
Chgb
|
chromogranin B
|
|
chr11_-_69419419
|
19.066
|
NM_013415
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr11_-_46125777
|
18.913
|
NM_133769
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2
|
|
chr19_-_45890238
|
18.776
|
NM_030716
NM_145703
NM_145704
|
Kcnip2
|
Kv channel-interacting protein 2
|
|
chr2_+_132607432
|
18.704
|
|
Chgb
|
chromogranin B
|
|
chr13_-_96214609
|
18.680
|
NM_198408
|
Crhbp
|
corticotropin releasing hormone binding protein
|
|
chr2_+_132607398
|
18.408
|
|
Chgb
|
chromogranin B
|
|
chr12_+_109572514
|
18.358
|
NM_010010
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1
|
|
chr13_-_58009599
|
18.250
|
|
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1
|
|
chr2_+_132607408
|
18.244
|
|
Chgb
|
chromogranin B
|
|
chr5_-_34630913
|
17.738
|
NM_001015039
|
Zfyve28
|
zinc finger, FYVE domain containing 28
|
|
chr16_-_67621054
|
17.541
|
NM_001145977
NM_178721
|
Cadm2
|
cell adhesion molecule 2
|
|
chr13_-_100286521
|
17.420
|
NM_008634
|
Mtap1b
|
microtubule-associated protein 1B
|
|
chr19_+_45635070
|
17.284
|
NM_172639
|
Dpcd
|
deleted in primary ciliary dyskinesia
|
|
chr17_+_35373506
|
17.117
|
NM_023179
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2
|
|
chr13_+_104899814
|
16.849
|
|
Sgtb
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta
|
|
chr18_-_43059156
|
16.847
|
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr1_-_166388431
|
16.694
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr9_-_21856445
|
16.016
|
NM_010487
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr5_-_68238444
|
15.723
|
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr9_+_103014463
|
15.669
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr6_-_23789259
|
15.643
|
NM_153163
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr14_-_57752270
|
15.114
|
NM_001010937
|
Gjb6
|
gap junction protein, beta 6
|
|
chr13_+_93195850
|
15.097
|
NM_175455
|
Ankrd34b
|
ankyrin repeat domain 34B
|
|
chrX_+_162676910
|
15.086
|
|
Gpm6b
|
glycoprotein m6b
|
|
chr6_+_4853319
|
15.022
|
NM_181595
|
Ppp1r9a
|
protein phosphatase 1, regulatory (inhibitor) subunit 9A
|
|
chr9_+_103014445
|
14.993
|
|
Rab6b
|
RAB6B, member RAS oncogene family
|
|
chr11_+_101017197
|
14.909
|
NM_134028
|
Tubg2
|
tubulin, gamma 2
|
|
chr9_+_120213190
|
14.808
|
NM_144557
|
Myrip
|
myosin VIIA and Rab interacting protein
|
|
chr2_-_73613381
|
14.664
|
NM_001113246
NM_001166603
|
Chn1
|
chimerin (chimaerin) 1
|
|
chr3_+_64913289
|
14.588
|
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr6_+_48487567
|
14.572
|
NM_133764
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr6_+_48487648
|
14.553
|
|
Atp6v0e2
|
ATPase, H+ transporting, lysosomal V0 subunit E2
|
|
chr9_-_42924626
|
14.374
|
NM_001034863
|
Tmem136
|
transmembrane protein 136
|
|
chr2_-_25174498
|
14.132
|
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1)
|
|
chr11_+_68505616
|
14.060
|
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle
|
|
chr2_+_70400179
|
14.051
|
NM_008077
|
Gad1
|
glutamic acid decarboxylase 1
|
|
chr1_+_78306890
|
14.010
|
NM_001004173
|
Sgpp2
|
sphingosine-1-phosphate phosphotase 2
|
|
chr3_+_145421870
|
14.009
|
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr1_-_89406861
|
14.001
|
NM_019867
|
Ngef
|
neuronal guanine nucleotide exchange factor
|
|
chr4_+_84851330
|
13.885
|
NM_019535
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr11_-_116274124
|
13.830
|
|
Rnf157
|
ring finger protein 157
|
|
chr2_+_25284190
|
13.820
|
NM_007379
|
Abca2
|
ATP-binding cassette, sub-family A (ABC1), member 2
|
|
chr4_-_119165183
|
13.707
|
NM_177572
|
Rimkla
|
ribosomal modification protein rimK-like family member A
|
|
chr11_+_68505612
|
13.486
|
|
Myh10
|
myosin, heavy polypeptide 10, non-muscle
|
|
chr13_-_100286446
|
13.420
|
|
Mtap1b
|
microtubule-associated protein 1B
|
|
chr17_-_57226987
|
13.412
|
NM_009451
|
Tubb4
|
tubulin, beta 4
|
|
chr18_-_31606908
|
13.399
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chr8_-_112692071
|
13.349
|
NM_007586
|
Calb2
|
calbindin 2
|
|
chr17_+_81343933
|
13.282
|
NM_026516
|
Tmem178
|
transmembrane protein 178
|
|
chr19_+_10525829
|
13.231
|
|
|
|
|
chr19_+_47089736
|
13.161
|
|
Ina
|
internexin neuronal intermediate filament protein, alpha
|
|
chr11_-_35793590
|
13.060
|
NM_170779
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1
|
|
chr14_-_57752102
|
12.917
|
|
Gjb6
|
gap junction protein, beta 6
|
|
chr1_+_34068669
|
12.881
|
NM_133833
NM_134448
|
Dst
|
dystonin
|
|
chr15_+_98465193
|
12.749
|
NM_007581
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr13_-_49243351
|
12.743
|
NM_029361
|
Wnk2
|
WNK lysine deficient protein kinase 2
|
|
chr18_+_38115192
|
12.601
|
NM_153793
|
Rell2
|
RELT-like 2
|
|
chr16_+_6349108
|
12.595
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr5_+_37637027
|
12.581
|
NM_007765
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr14_+_104049411
|
12.486
|
|
Slain1
|
SLAIN motif family, member 1
|
|
chr2_-_121632749
|
12.475
|
NM_172673
|
Frmd5
|
FERM domain containing 5
|
|
chr12_+_91050912
|
12.362
|
|
Nrxn3
|
neurexin III
|
|
chr2_-_25174662
|
12.354
|
NM_001177656
NM_001177657
NM_008169
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1)
|
|
chr9_-_75531734
|
12.314
|
NM_001164790
NM_009130
|
Scg3
|
secretogranin III
|
|
chr6_-_77929575
|
12.303
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr11_+_71564481
|
12.283
|
|
Wscd1
|
WSC domain containing 1
|
|
chr12_+_113960424
|
12.270
|
|
AW555464
|
expressed sequence AW555464
|
|
chrX_+_155852439
|
12.206
|
NM_001081124
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr17_+_27792487
|
12.164
|
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr2_+_90729768
|
12.116
|
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr17_+_27792587
|
12.113
|
NM_011861
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1
|
|
chr11_-_115229039
|
12.051
|
|
C630004H02Rik
|
RIKEN cDNA C630004H02 gene
|
|
chr8_-_4217275
|
11.968
|
NM_207203
|
BC068157
|
cDNA sequence BC068157
|
|
chr8_-_4259126
|
11.934
|
NM_183315
|
Ctxn1
|
cortexin 1
|
|
chr1_-_84692352
|
11.913
|
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chr5_+_37259906
|
11.911
|
|
Ppp2r2c
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), gamma isoform
|
|
chr12_+_113960530
|
11.906
|
|
AW555464
|
expressed sequence AW555464
|
|
chr3_+_145421621
|
11.885
|
NM_026993
|
Ddah1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr11_+_42233258
|
11.882
|
NM_008070
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr7_+_4071087
|
11.694
|
NM_001001454
NM_001109765
NM_021324
|
Ttyh1
|
tweety homolog 1 (Drosophila)
|
|
chr7_-_30066642
|
11.670
|
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr5_+_25265761
|
11.588
|
NM_001004365
|
Actr3b
|
ARP3 actin-related protein 3 homolog B (yeast)
|
|
chr10_-_80945379
|
11.547
|
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr8_-_128422192
|
11.383
|
NM_175561
|
Pcnxl2
|
pecanex-like 2 (Drosophila)
|
|
chr3_+_156224757
|
11.338
|
NM_001039094
NM_177274
|
Negr1
|
neuronal growth regulator 1
|
|
chr4_+_101169252
|
11.311
|
NM_198412
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr5_-_68238654
|
11.102
|
NM_001038999
NM_009727
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1
|
|
chr13_-_58009653
|
11.083
|
NM_001166463
NM_001166464
NM_001166465
NM_001166466
NM_009262
|
Spock1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 1
|
|
chr16_-_34263272
|
11.082
|
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr14_-_119324535
|
11.033
|
|
Dzip1
|
DAZ interacting protein 1
|
|
chr4_+_108551674
|
11.008
|
NM_023537
|
Rab3b
|
RAB3B, member RAS oncogene family
|
|
chr9_+_107838164
|
10.905
|
NM_145621
|
Camkv
|
CaM kinase-like vesicle-associated
|
|
chr7_+_129432810
|
10.891
|
|
Prkcb
|
protein kinase C, beta
|
|
chr4_+_84851440
|
10.860
|
|
Sh3gl2
|
SH3-domain GRB2-like 2
|
|
chr13_+_58909254
|
10.845
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr4_+_15808409
|
10.802
|
NM_009788
|
Calb1
|
calbindin 1
|
|
chr13_+_58909648
|
10.686
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr5_+_37259780
|
10.683
|
NM_172994
|
Ppp2r2c
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), gamma isoform
|
|
chr4_+_137263376
|
10.618
|
|
Rap1gap
|
Rap1 GTPase-activating protein
|
|
chr13_-_25029458
|
10.478
|
NM_172532
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1
|
|
chr1_+_179372452
|
10.475
|
|
Zfp238
|
zinc finger protein 238
|
|
chr1_+_74379182
|
10.472
|
NM_019999
|
Pnkd
|
paroxysmal nonkinesiogenic dyskinesia
|
|
chr6_+_114232618
|
10.464
|
NM_178703
|
Slc6a1
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 1
|
|
chr6_-_8728094
|
10.457
|
|
Ica1
|
islet cell autoantigen 1
|
|
chr4_+_129662110
|
10.402
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr13_-_92901406
|
10.376
|
NM_009027
|
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2
|
|
chr7_-_31230577
|
10.374
|
NM_007467
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr3_+_108163704
|
10.333
|
|
Sort1
|
sortilin 1
|
|
chr9_+_75161942
|
10.216
|
NM_138719
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr11_-_74403603
|
10.204
|
NM_001015046
|
Rap1gap2
|
RAP1 GTPase activating protein 2
|
|
chr6_-_126658429
|
10.178
|
|
|
|
|
chr10_-_80945445
|
10.162
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr8_-_97220127
|
10.139
|
NM_026385
|
Pllp
|
plasma membrane proteolipid
|
|
chr7_-_30066867
|
10.136
|
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chr14_-_62911797
|
10.089
|
NM_173419
|
Dleu7
|
deleted in lymphocytic leukemia, 7
|
|
chr7_+_25266986
|
10.075
|
NM_153112
|
Cadm4
|
cell adhesion molecule 4
|
|
chr16_-_34263047
|
10.074
|
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr7_-_31230463
|
10.069
|
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr9_-_86775282
|
10.044
|
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr4_-_135050358
|
9.951
|
NM_028995
|
Nipal3
|
NIPA-like domain containing 3
|
|
chr3_-_88576415
|
9.897
|
NM_018804
|
Syt11
|
synaptotagmin XI
|
|
chr11_+_101017307
|
9.872
|
|
Tubg2
|
tubulin, gamma 2
|
|
chr7_-_30066910
|
9.842
|
NM_001082548
NM_011464
|
Spint2
|
serine protease inhibitor, Kunitz type 2
|
|
chrX_+_71550562
|
9.828
|
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1
|
|
chr6_+_21166108
|
9.818
|
NM_019697
|
Kcnd2
|
potassium voltage-gated channel, Shal-related family, member 2
|
|
chr15_-_84388113
|
9.800
|
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr3_+_64913564
|
9.769
|
NM_010597
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1
|
|
chr15_-_84387921
|
9.738
|
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like
|
|
chr12_-_100139666
|
9.706
|
NM_001081191
|
Eml5
|
echinoderm microtubule associated protein like 5
|
|
chrX_-_131543723
|
9.681
|
NM_175446
|
Zmat1
|
zinc finger, matrin type 1
|
|
chr16_+_91270319
|
9.617
|
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr13_+_58909236
|
9.612
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr10_-_80306743
|
9.589
|
NM_001013758
|
Lingo3
|
leucine rich repeat and Ig domain containing 3
|
|
chr7_+_134094045
|
9.550
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr8_+_4493404
|
9.542
|
NM_026058
|
Lass4
|
LAG1 homolog, ceramide synthase 4
|
|
chr15_-_33617572
|
9.502
|
NM_001085421
|
Tspyl5
|
testis-specific protein, Y-encoded-like 5
|
|
chr7_-_31230564
|
9.488
|
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr8_+_15032567
|
9.488
|
|
Kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11
|
|
chr13_+_58909168
|
9.472
|
NM_001025074
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr12_+_16660521
|
9.458
|
|
Ntsr2
|
neurotensin receptor 2
|
|
chr15_-_37721754
|
9.452
|
NM_001170866
|
Ncald
|
neurocalcin delta
|
|
chr15_-_37722161
|
9.415
|
NM_134094
|
Ncald
|
neurocalcin delta
|
|
chr17_-_26338254
|
9.414
|
NM_008113
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma
|
|
chr10_-_32609590
|
9.410
|
NM_001013411
NM_001025286
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2
|
|
chrX_-_70905384
|
9.409
|
|
Pnck
|
pregnancy upregulated non-ubiquitously expressed CaM kinase
|
|
chr8_+_112603609
|
9.387
|
NM_175480
|
Zfp612
|
zinc finger protein 612
|
|
chr2_-_170232008
|
9.385
|
|
Bcas1
|
breast carcinoma amplified sequence 1
|
|
chr7_+_4874478
|
9.367
|
|
Nat14
|
N-acetyltransferase 14
|
|
chr8_+_4493451
|
9.306
|
|
Lass4
|
LAG1 homolog, ceramide synthase 4
|
|
chr15_+_79059802
|
9.242
|
NM_001045558
|
Pick1
|
protein interacting with C kinase 1
|
|
chr14_-_122196929
|
9.235
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr7_+_133416177
|
9.193
|
|
Sbk1
|
SH3-binding kinase 1
|
|
chr14_-_71035662
|
9.123
|
|
Epb4.9
|
erythrocyte protein band 4.9
|
|
chr15_-_64753721
|
9.112
|
NM_009623
|
Adcy8
|
adenylate cyclase 8
|
|
chr9_-_86775154
|
9.099
|
NM_013669
|
Snap91
|
synaptosomal-associated protein 91
|
|
chr16_+_7069927
|
9.086
|
NM_183188
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr17_-_26338106
|
9.081
|
|
Arhgdig
|
Rho GDP dissociation inhibitor (GDI) gamma
|