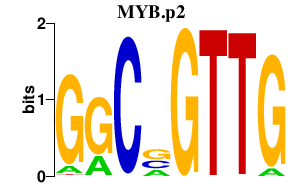
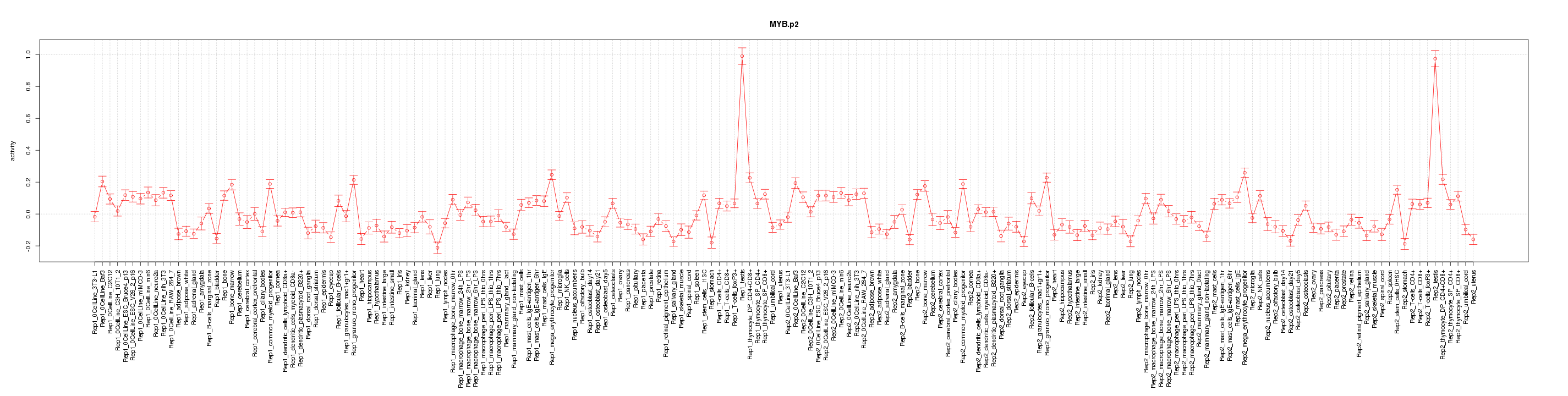
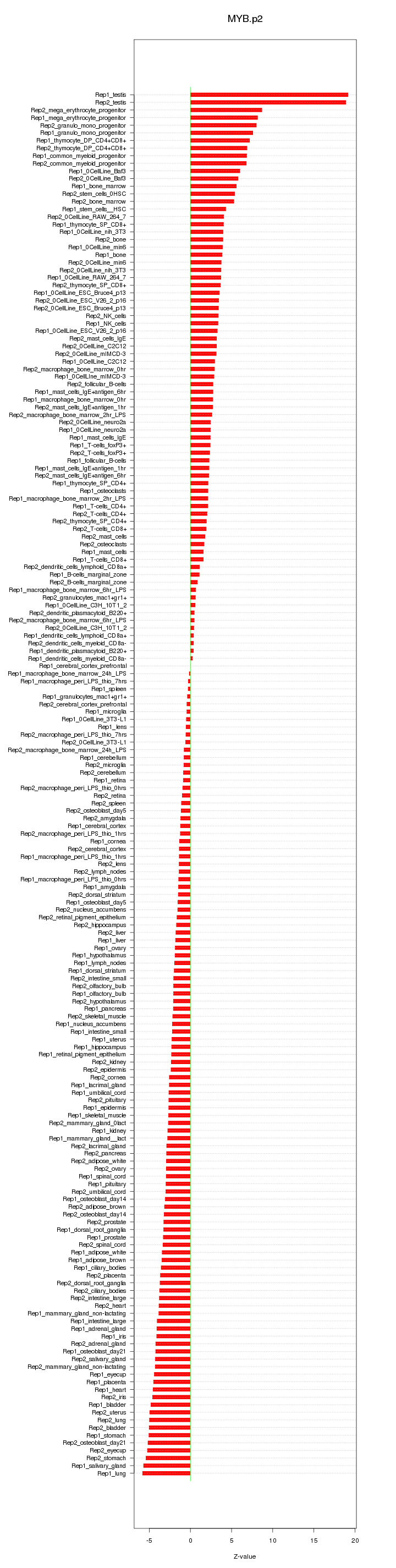
|
chr4_-_83132289
|
46.315
|
NM_133948
|
Psip1
|
PC4 and SFRS1 interacting protein 1
|
|
chr1_-_171461510
|
38.032
|
NM_023284
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr6_+_116214215
|
36.917
|
NM_001081317
|
Anubl1
|
AN1, ubiquitin-like, homolog (Xenopus laevis)
|
|
chr2_-_37558794
|
36.548
|
|
Strbp
|
spermatid perinuclear RNA binding protein
|
|
chr3_+_40549303
|
36.509
|
|
Hspa4l
|
heat shock protein 4 like
|
|
chr14_-_32064403
|
35.746
|
NM_027949
|
Phf7
|
PHD finger protein 7
|
|
chr2_-_172195998
|
35.068
|
NM_011497
|
Aurka
|
aurora kinase A
|
|
chr9_+_72286335
|
34.232
|
NM_008613
|
Mns1
|
meiosis-specific nuclear structural protein 1
|
|
chr4_-_118109775
|
34.209
|
NM_023223
|
Cdc20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chr8_+_86239094
|
33.880
|
NM_197982
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39
|
|
chr18_-_46685557
|
31.960
|
NM_173423
|
Fem1c
|
fem-1 homolog c (C.elegans)
|
|
chr19_-_9210094
|
31.876
|
|
Asrgl1
|
asparaginase like 1
|
|
chr2_+_118872854
|
31.803
|
NM_029617
|
Casc5
|
cancer susceptibility candidate 5
|
|
chr18_-_46685512
|
31.043
|
|
Fem1c
|
fem-1 homolog c (C.elegans)
|
|
chr4_-_124614072
|
30.844
|
NM_026560
|
Cdca8
|
cell division cycle associated 8
|
|
chr14_+_47380215
|
30.756
|
NM_028222
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr1_-_133034721
|
30.495
|
NM_145508
|
Dyrk3
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3
|
|
chr19_-_9210047
|
29.791
|
NM_025610
|
Asrgl1
|
asparaginase like 1
|
|
chr8_+_85913750
|
28.949
|
NM_009904
|
Clgn
|
calmegin
|
|
chr14_-_66696533
|
28.731
|
NM_009618
|
Adam2
|
a disintegrin and metallopeptidase domain 2
|
|
chr14_+_47380287
|
28.611
|
|
Cdkn3
|
cyclin-dependent kinase inhibitor 3
|
|
chr2_+_17986187
|
28.339
|
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10
|
|
chr17_+_8358382
|
28.314
|
NM_001197046
NM_201230
|
Fgfr1op
|
Fgfr1 oncogene partner
|
|
chr16_-_16073490
|
28.002
|
|
2310008H04Rik
|
RIKEN cDNA 2310008H04 gene
|
|
chr18_+_34784277
|
27.952
|
NM_001166407
|
Kif20a
|
kinesin family member 20A
|
|
chr1_-_193399412
|
27.841
|
NM_029766
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr5_-_31522889
|
27.722
|
NM_008014
|
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform
|
|
chr8_+_86239131
|
27.692
|
|
Ddx39
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 39
|
|
chr9_+_106183391
|
27.120
|
NM_027354
|
Poc1a
|
POC1 centriolar protein homolog A (Chlamydomonas)
|
|
chr2_-_127657532
|
26.909
|
NM_001113179
NM_009772
|
Bub1
|
budding uninhibited by benzimidazoles 1 homolog (S. cerevisiae)
|
|
chr7_+_134005943
|
26.805
|
|
Hirip3
|
HIRA interacting protein 3
|
|
chr14_+_70240213
|
25.912
|
NM_028526
NM_028560
|
Pebp4
|
phosphatidylethanolamine binding protein 4
|
|
chr11_+_69749297
|
25.776
|
NM_016875
|
Ybx2
|
Y box protein 2
|
|
chr1_-_44159187
|
25.752
|
NM_029368
|
1700029F09Rik
|
RIKEN cDNA 1700029F09 gene
|
|
chr3_+_116297899
|
25.714
|
NM_028349
|
Sass6
|
spindle assembly 6 homolog (C. elegans)
|
|
chrX_+_130842622
|
25.623
|
|
Cenpi
|
centromere protein I
|
|
chr2_-_126959605
|
25.622
|
NM_144818
|
Ncaph
|
non-SMC condensin I complex, subunit H
|
|
chr5_+_124199713
|
25.535
|
NM_001042421
|
Kntc1
|
kinetochore associated 1
|
|
chr17_-_71200462
|
25.346
|
NM_009372
|
Tgif1
|
TGFB-induced factor homeobox 1
|
|
chr6_+_145564482
|
25.230
|
NM_009449
|
Tuba3b
|
tubulin, alpha 3B
|
|
chrX_+_130842701
|
25.205
|
NM_145924
|
Cenpi
|
centromere protein I
|
|
chr18_+_34784589
|
24.816
|
NM_009004
NM_001166406
|
Kif20a
|
kinesin family member 20A
|
|
chr17_+_31701700
|
24.752
|
NM_016670
|
Pknox1
|
Pbx/knotted 1 homeobox
|
|
chr8_+_86479405
|
24.727
|
NM_024184
|
Asf1b
|
ASF1 anti-silencing function 1 homolog B (S. cerevisiae)
|
|
chr11_+_23565673
|
24.491
|
NM_001033654
NM_028956
|
Pus10
|
pseudouridylate synthase 10
|
|
chr10_-_22451738
|
24.109
|
|
Tbpl1
|
TATA box binding protein-like 1
|
|
chr3_+_40549475
|
23.914
|
NM_011020
|
Hspa4l
|
heat shock protein 4 like
|
|
chr11_+_23565975
|
23.206
|
NM_028304
|
Pus10
|
pseudouridylate synthase 10
|
|
chr17_-_7165497
|
23.049
|
NM_147155
|
Tagap1
|
T-cell activation GTPase activating protein 1
|
|
chr6_-_125141525
|
22.957
|
NM_146171
|
Ncapd2
|
non-SMC condensin I complex, subunit D2
|
|
chr4_+_140258090
|
22.612
|
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr10_+_127114119
|
21.986
|
NM_001113211
NM_173732
|
Tmem194
|
transmembrane protein 194
|
|
chr5_+_135625533
|
21.811
|
NM_013763
|
Tbl2
|
transducin (beta)-like 2
|
|
chr4_-_44180323
|
21.706
|
NM_175201
|
Rnf38
|
ring finger protein 38
|
|
chr13_-_26861883
|
21.290
|
NM_008232
|
Hdgfl1
|
hepatoma derived growth factor-like 1
|
|
chr8_+_106774774
|
21.282
|
NM_029295
NM_001037841
|
Cklf
|
chemokine-like factor
|
|
chr5_+_38611707
|
21.263
|
NM_025281
|
Lyar
|
Ly1 antibody reactive clone
|
|
chr11_+_87430665
|
21.251
|
NM_001045527
|
Hsf5
|
heat shock transcription factor family member 5
|
|
chr2_+_118639930
|
21.229
|
|
D2Ertd750e
|
DNA segment, Chr 2, ERATO Doi 750, expressed
|
|
chr12_-_80292805
|
21.220
|
NM_021557
|
Rdh11
|
retinol dehydrogenase 11
|
|
chr2_+_118639738
|
20.996
|
NM_026412
|
D2Ertd750e
|
DNA segment, Chr 2, ERATO Doi 750, expressed
|
|
chr1_+_58052814
|
20.405
|
NM_199007
|
Sgol2
|
shugoshin-like 2 (S. pombe)
|
|
chr5_-_8422533
|
20.212
|
NM_001190717
NM_013726
|
Dbf4
|
DBF4 homolog (S. cerevisiae)
|
|
chr17_-_34027575
|
19.371
|
NM_001195298
|
Kifc1
|
kinesin family member C1
|
|
chr11_+_105043195
|
19.302
|
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis)
|
|
chr11_+_102526898
|
19.134
|
NM_001127576
|
Gm1564
|
predicted gene 1564
|
|
chr9_+_20833816
|
18.648
|
NM_023892
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group
|
|
chr17_-_34027488
|
18.509
|
|
Kifc1
|
kinesin family member C1
|
|
chr1_-_193399352
|
18.392
|
|
Dtl
|
denticleless homolog (Drosophila)
|
|
chr2_-_164860272
|
18.239
|
NM_144892
|
Ncoa5
|
nuclear receptor coactivator 5
|
|
chr12_-_80292830
|
18.229
|
|
Rdh11
|
retinol dehydrogenase 11
|
|
chr11_+_83116262
|
18.196
|
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr10_-_119913990
|
18.101
|
NM_010441
|
Hmga2
|
high mobility group AT-hook 2
|
|
chr6_-_146902928
|
17.940
|
NM_001164236
NM_028509
|
1700034J05Rik
|
RIKEN cDNA 1700034J05 gene
|
|
chr2_-_144156799
|
17.579
|
|
Ovol2
|
ovo-like 2 (Drosophila)
|
|
chr7_-_52689639
|
17.578
|
|
Ruvbl2
|
RuvB-like protein 2
|
|
chr4_+_129168433
|
17.515
|
NM_001085491
|
1700125D06Rik
|
RIKEN cDNA 1700125D06 gene
|
|
chr18_+_56867378
|
17.351
|
NM_010721
|
Lmnb1
|
lamin B1
|
|
chr11_-_105042428
|
17.294
|
|
1700052K11Rik
|
RIKEN cDNA 1700052K11 gene
|
|
chr19_-_29399819
|
17.152
|
|
Insl6
|
insulin-like 6
|
|
chr1_-_168168870
|
17.110
|
NM_175296
|
Mael
|
maelstrom homolog (Drosophila)
|
|
chr12_+_66326503
|
17.064
|
NM_027614
|
Wdr20b
|
WD repeat domain 20b
|
|
chr5_-_31522802
|
16.968
|
|
Ppm1g
|
protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform
|
|
chr17_-_34027547
|
16.898
|
|
Kifc1
|
kinesin family member C1
|
|
chr6_+_8459595
|
16.836
|
NM_133236
|
Glcci1
|
glucocorticoid induced transcript 1
|
|
chr1_-_158233462
|
16.812
|
NM_001134741
|
Tdrd5
|
tudor domain containing 5
|
|
chr6_-_88834347
|
16.731
|
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae)
|
|
chr2_-_121239527
|
16.539
|
NM_153075
|
Catsper2
|
cation channel, sperm associated 2
|
|
chr7_-_17509228
|
16.523
|
NM_007590
|
Calm3
|
calmodulin 3
|
|
chr11_+_62875160
|
16.362
|
NM_027660
|
Tekt3
|
tektin 3
|
|
chr14_-_87540995
|
16.218
|
|
Diap3
|
diaphanous homolog 3 (Drosophila)
|
|
chr6_-_28211600
|
16.159
|
NM_001081678
|
Zfp800
|
zinc finger protein 800
|
|
chr19_-_29399804
|
16.150
|
NM_013754
|
Insl6
|
insulin-like 6
|
|
chr5_+_34001199
|
16.143
|
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3
|
|
chr12_-_92622769
|
16.137
|
NM_181815
|
4930534B04Rik
|
RIKEN cDNA 4930534B04 gene
|
|
chr6_-_28211910
|
16.015
|
|
Zfp800
|
zinc finger protein 800
|
|
chr1_-_158869650
|
15.984
|
NM_001159965
NM_001159967
NM_001159968
NM_023884
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr2_-_144157097
|
15.892
|
NM_152947
|
Ovol2
|
ovo-like 2 (Drosophila)
|
|
chr5_+_3657003
|
15.781
|
NM_029141
NM_029154
|
4930511M11Rik
|
RIKEN cDNA 4930511M11 gene
|
|
chr10_+_79689342
|
15.745
|
NM_023431
|
Mum1
|
melanoma associated antigen (mutated) 1
|
|
chr7_+_13610047
|
15.741
|
|
Trim28
|
tripartite motif-containing 28
|
|
chr3_+_146067763
|
15.593
|
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr8_-_25836259
|
15.587
|
NM_009619
|
Adam3
|
a disintegrin and metallopeptidase domain 3 (cyritestin)
|
|
chr7_+_134005485
|
15.540
|
NM_172746
|
Hirip3
|
HIRA interacting protein 3
|
|
chrX_+_97821403
|
15.482
|
NM_008446
|
Kif4
|
kinesin family member 4
|
|
chr2_-_3336293
|
15.242
|
|
Meig1
|
meiosis expressed gene 1
|
|
chr3_+_146067605
|
15.158
|
NM_138744
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr6_+_136467388
|
15.111
|
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr9_+_107449628
|
15.108
|
NM_053253
|
Zmynd10
|
zinc finger, MYND domain containing 10
|
|
chr9_-_35365985
|
15.097
|
NM_013932
|
Ddx25
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 25
|
|
chr7_+_52071438
|
15.026
|
|
Nup62
|
nucleoporin 62
|
|
chr11_-_33413625
|
14.921
|
|
Ranbp17
|
RAN binding protein 17
|
|
chr3_-_51144470
|
14.829
|
|
Elf2
|
E74-like factor 2
|
|
chr1_-_158869401
|
14.710
|
NM_001159966
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2
|
|
chr1_+_193645351
|
14.694
|
NM_010892
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr15_-_103260834
|
14.638
|
NM_028797
|
Gtsf1
|
gametocyte specific factor 1
|
|
chr8_+_73118329
|
14.624
|
NM_023627
|
Isyna1
|
myo-inositol 1-phosphate synthase A1
|
|
chr6_+_89594017
|
14.617
|
|
Txnrd3
|
thioredoxin reductase 3
|
|
chr7_+_52071622
|
14.534
|
NM_001171024
NM_053074
|
Nup62-il4i1
Nup62
|
Nup62-Il4i1 protein
nucleoporin 62
|
|
chrX_+_20002809
|
14.517
|
|
Phf16
|
PHD finger protein 16
|
|
chr11_+_62660624
|
14.499
|
NM_001033669
|
Fbxw10
|
F-box and WD-40 domain protein 10
|
|
chr4_-_43512476
|
14.490
|
NM_001013377
|
E130306D19Rik
|
RIKEN cDNA E130306D19 gene
|
|
chr6_+_54766562
|
14.460
|
NM_199143
|
Znrf2
|
zinc and ring finger 2
|
|
chr10_-_88906629
|
14.430
|
NM_001033331
NM_001079876
|
Gas2l3
|
growth arrest-specific 2 like 3
|
|
chr19_+_36484237
|
14.386
|
|
Pcgf5
|
polycomb group ring finger 5
|
|
chr17_+_47736273
|
14.255
|
|
Ccnd3
|
cyclin D3
|
|
chr2_+_122463622
|
14.218
|
NM_001004174
|
AA467197
|
expressed sequence AA467197
|
|
chr7_-_52689833
|
14.191
|
NM_011304
|
Ruvbl2
|
RuvB-like protein 2
|
|
chr14_+_56364551
|
14.153
|
|
2610027L16Rik
|
RIKEN cDNA 2610027L16 gene
|
|
chr6_+_89593981
|
14.089
|
NM_001178058
NM_001178059
NM_001178060
NM_153162
|
Txnrd3
|
thioredoxin reductase 3
|
|
chr7_-_104886507
|
14.074
|
|
Aqp11
|
aquaporin 11
|
|
chr14_+_56364519
|
14.071
|
NM_026403
|
2610027L16Rik
|
RIKEN cDNA 2610027L16 gene
|
|
chr3_+_137527448
|
14.051
|
NM_016750
|
H2afz
|
H2A histone family, member Z
|
|
chr11_+_100575163
|
14.042
|
NM_029307
|
Hspb9
|
heat shock protein, alpha-crystallin-related, B9
|
|
chr7_+_29225955
|
14.021
|
NM_001039192
|
Gmfg
|
glia maturation factor, gamma
|
|
chr3_+_40512792
|
14.014
|
NM_178386
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chr2_-_164860303
|
13.975
|
|
Ncoa5
|
nuclear receptor coactivator 5
|
|
chr2_-_119303348
|
13.863
|
NM_026574
|
Ino80
|
INO80 homolog (S. cerevisiae)
|
|
chr6_-_113327472
|
13.860
|
NM_133932
|
Tada3
|
transcriptional adaptor 3
|
|
chr14_+_35487052
|
13.804
|
NM_001004436
|
Wapal
|
wings apart-like homolog (Drosophila)
|
|
chr4_-_107356766
|
13.761
|
NM_019872
|
Dmrtb1
|
DMRT-like family B with proline-rich C-terminal, 1
|
|
chr13_+_22073049
|
13.737
|
NM_001162928
NM_001162929
|
Pom121l2
|
POM121 membrane glycoprotein-like 2 (rat)
|
|
chr11_+_105043224
|
13.723
|
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis)
|
|
chr4_-_116855158
|
13.539
|
NM_134471
|
Kif2c
|
kinesin family member 2C
|
|
chr1_-_163806710
|
13.509
|
NM_172645
|
AI848100
|
expressed sequence AI848100
|
|
chr5_-_123439064
|
13.473
|
NM_001003953
|
Kdm2b
|
lysine (K)-specific demethylase 2B
|
|
chr18_-_70689894
|
13.470
|
NM_011972
|
Poli
|
polymerase (DNA directed), iota
|
|
chr9_+_108295164
|
13.420
|
NM_053216
|
1700102P08Rik
|
RIKEN cDNA 1700102P08 gene
|
|
chr3_-_68930721
|
13.391
|
|
Kpna4
|
karyopherin (importin) alpha 4
|
|
chr1_+_75139300
|
13.350
|
NM_170755
|
Fam134a
|
family with sequence similarity 134, member A
|
|
chr15_+_4848736
|
13.341
|
NM_001166066
|
Heatr7b2
|
XVHEAT repeat family member 7B2
|
|
chr16_+_36935029
|
13.333
|
NM_008225
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1
|
|
chr19_-_41876509
|
13.296
|
NM_001163495
NM_027667
|
Arhgap19
|
Rho GTPase activating protein 19
|
|
chr6_+_136467315
|
13.291
|
NM_019426
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr10_-_22451093
|
13.240
|
|
Tbpl1
|
TATA box binding protein-like 1
|
|
chr6_-_131338465
|
13.229
|
NM_011733
NM_139117
|
Csda
|
cold shock domain protein A
|
|
chr1_+_40497472
|
13.213
|
NM_001025602
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr1_-_60155312
|
13.203
|
NM_001199060
|
Wdr12
|
WD repeat domain 12
|
|
chr4_-_116300533
|
13.160
|
NM_016777
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding)
|
|
chr18_-_25327116
|
12.945
|
|
5730494M16Rik
|
RIKEN cDNA 5730494M16 gene
|
|
chr3_+_137527575
|
12.838
|
|
H2afz
|
H2A histone family, member Z
|
|
chr12_+_34000269
|
12.828
|
|
Atxn7l1
|
ataxin 7-like 1
|
|
chr12_-_70299271
|
12.809
|
NM_027269
|
1110034A24Rik
|
RIKEN cDNA 1110034A24 gene
|
|
chr15_-_36538484
|
12.806
|
|
Pabpc1
|
poly(A) binding protein, cytoplasmic 1
|
|
chr11_-_82577789
|
12.778
|
NM_009839
|
Cct6b
|
chaperonin containing Tcp1, subunit 6b (zeta)
|
|
chr9_+_30838224
|
12.727
|
NM_001115130
NM_172765
|
Zbtb44
|
zinc finger and BTB domain containing 44
|
|
chr5_-_125342474
|
12.666
|
NM_144819
|
Ccdc92
|
coiled-coil domain containing 92
|
|
chr7_-_134169938
|
12.632
|
NM_010772
|
Maz
|
MYC-associated zinc finger protein (purine-binding transcription factor)
|
|
chrX_-_99352341
|
12.626
|
NM_146235
|
Ercc6l
|
excision repair cross-complementing rodent repair deficiency complementation group 6 - like
|
|
chr8_-_25785199
|
12.623
|
NM_010084
|
Adam18
|
a disintegrin and metallopeptidase domain 18
|
|
chr14_-_63379948
|
12.581
|
NM_008715
|
Ints6
|
integrator complex subunit 6
|
|
chr2_+_172196428
|
12.555
|
NM_024199
|
Cstf1
|
cleavage stimulation factor, 3' pre-RNA, subunit 1
|
|
chr8_-_75072164
|
12.529
|
NM_027485
|
Med26
|
mediator complex subunit 26
|
|
chr17_-_71876127
|
12.525
|
|
Ndc80
|
NDC80 homolog, kinetochore complex component (S. cerevisiae)
|
|
chr5_+_7304126
|
12.525
|
NM_027603
|
4921511H03Rik
|
RIKEN cDNA 4921511H03 gene
|
|
chr6_-_113327369
|
12.492
|
|
Tada3
|
transcriptional adaptor 3
|
|
chr1_-_55419550
|
12.486
|
NM_001113367
|
Boll
|
bol, boule-like (Drosophila)
|
|
chr12_-_85489188
|
12.472
|
NM_027438
|
Pnma1
|
paraneoplastic antigen MA1
|
|
chr15_-_83749802
|
12.456
|
NM_001161629
|
Efcab6
|
EF-hand calcium binding domain 6
|
|
chr18_-_34911152
|
12.454
|
NM_009860
|
Cdc25c
|
cell division cycle 25 homolog C (S. pombe)
|
|
chr11_+_23156482
|
12.438
|
|
Xpo1
|
exportin 1, CRM1 homolog (yeast)
|
|
chr15_+_57743727
|
12.437
|
NM_001081396
NM_001167679
|
Wdr67
|
WD repeat domain 67
|
|
chr14_-_99498905
|
12.428
|
NM_028315
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae)
|
|
chr17_-_9862538
|
12.406
|
NM_001163836
|
Pabpc6
|
poly(A) binding protein, cytoplasmic 6
|
|
chr2_+_53050461
|
12.243
|
|
Arl6ip6
|
ADP-ribosylation factor-like 6 interacting protein 6
|
|
chr2_-_72817311
|
12.219
|
NM_001098425
|
Sp3
|
trans-acting transcription factor 3
|
|
chr8_+_86132073
|
12.213
|
NM_018808
|
Dnajb1
|
DnaJ (Hsp40) homolog, subfamily B, member 1
|
|
chr11_+_97223851
|
12.184
|
NM_138657
|
Socs7
|
suppressor of cytokine signaling 7
|
|
chr15_+_89398756
|
12.115
|
NM_013455
|
Acr
|
acrosin prepropeptide
|
|
chr9_-_62129985
|
12.081
|
NM_025721
|
Spesp1
|
sperm equatorial segment protein 1
|
|
chr11_-_78364173
|
12.038
|
NM_133706
|
Tmem97
|
transmembrane protein 97
|
|
chr13_-_101556356
|
12.030
|
NM_172301
|
Ccnb1
|
cyclin B1
|
|
chrX_+_20002875
|
12.027
|
|
Phf16
|
PHD finger protein 16
|
|
chr3_+_40512853
|
11.983
|
|
Slc25a31
|
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31
|
|
chr1_-_75139207
|
11.903
|
|
1810031K17Rik
|
RIKEN cDNA 1810031K17 gene
|
|
chr18_-_70689871
|
11.798
|
|
Poli
|
polymerase (DNA directed), iota
|
|
chr7_+_135056028
|
11.788
|
NM_026370
|
Myst1
|
MYST histone acetyltransferase 1
|
|
chr2_-_25127860
|
11.783
|
NM_023464
|
Ssna1
|
Sjogren's syndrome nuclear autoantigen 1
|
|
chr2_+_125977876
|
11.747
|
NM_026981
|
Dtwd1
|
DTW domain containing 1
|
|
chr11_-_68740550
|
11.731
|
NM_145746
|
Odf4
|
outer dense fiber of sperm tails 4
|