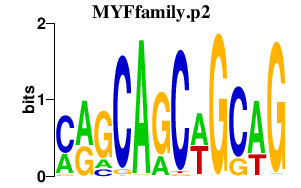
|
chrX_-_119510890
|
63.551
|
NM_138742
|
Nap1l3
|
nucleosome assembly protein 1-like 3
|
|
chr8_-_9771020
|
52.359
|
NM_173446
|
Fam155a
|
family with sequence similarity 155, member A
|
|
chr9_-_112088452
|
40.676
|
NM_001177618
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr2_+_83652846
|
35.894
|
NM_175514
|
Fam171b
|
family with sequence similarity 171, member B
|
|
chr4_+_104829951
|
30.944
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr18_+_45719732
|
29.479
|
NM_080465
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr4_+_104829963
|
28.668
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr9_+_75473524
|
27.256
|
NM_027309
|
Lysmd2
|
LysM, putative peptidoglycan-binding, domain containing 2
|
|
chr2_+_48805027
|
26.314
|
NM_029924
|
Mbd5
|
methyl-CpG binding domain protein 5
|
|
chr12_-_73509668
|
25.503
|
|
Rtn1
|
reticulon 1
|
|
chr6_-_77929543
|
24.716
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr2_+_157387284
|
23.863
|
|
Nnat
|
neuronatin
|
|
chr4_+_104830022
|
23.681
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr18_+_45719464
|
23.458
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr6_-_77929685
|
23.375
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr17_+_35373506
|
23.325
|
NM_023179
|
Atp6v1g2
|
ATPase, H+ transporting, lysosomal V1 subunit G2
|
|
chr12_-_71448600
|
23.250
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr9_+_27598853
|
23.020
|
NM_177906
|
Opcml
|
opioid binding protein/cell adhesion molecule-like
|
|
chr6_-_126690651
|
22.792
|
NM_013568
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6
|
|
chr12_-_73509537
|
22.481
|
|
Rtn1
|
reticulon 1
|
|
chr18_+_45719101
|
21.411
|
|
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2
|
|
chr18_-_43058544
|
20.940
|
NM_027531
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr13_+_64263131
|
20.486
|
NM_019986
|
Habp4
|
hyaluronic acid binding protein 4
|
|
chr16_-_67621054
|
19.536
|
NM_001145977
NM_178721
|
Cadm2
|
cell adhesion molecule 2
|
|
chr13_-_69877709
|
19.105
|
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr1_-_125942073
|
18.631
|
NM_199021
|
Dpp10
|
dipeptidylpeptidase 10
|
|
chrX_-_131745187
|
18.392
|
NM_025355
|
Tceal6
|
transcription elongation factor A (SII)-like 6
|
|
chr16_+_6349108
|
17.803
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr1_-_84692352
|
17.735
|
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chrX_-_100381973
|
17.613
|
NM_008671
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr6_-_77929660
|
17.608
|
NM_001109764
NM_009819
NM_145732
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr13_+_58909236
|
17.382
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr12_-_71448300
|
17.241
|
|
Trim9
|
tripartite motif-containing 9
|
|
chr18_-_43847367
|
17.162
|
NM_001163637
|
Jakmip2
|
janus kinase and microtubule interacting protein 2
|
|
chr16_-_28753276
|
17.075
|
NM_010199
|
Fgf12
|
fibroblast growth factor 12
|
|
chr13_+_58909648
|
16.251
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr5_-_38550627
|
15.934
|
NM_010942
|
Nsg1
|
neuron specific gene family member 1
|
|
chr6_-_23789048
|
15.754
|
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr12_-_73509925
|
15.379
|
NM_153457
|
Rtn1
|
reticulon 1
|
|
chr8_+_128519295
|
15.336
|
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr13_-_78337844
|
15.225
|
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr4_+_104829867
|
15.153
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr13_+_58909254
|
14.694
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr9_+_45178267
|
14.631
|
NM_022004
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6
|
|
chr7_+_138079669
|
14.549
|
NM_019564
|
Htra1
|
HtrA serine peptidase 1
|
|
chr2_-_84614976
|
14.544
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr9_+_45178485
|
14.482
|
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6
|
|
chr16_-_4419586
|
14.292
|
NM_009624
|
Adcy9
|
adenylate cyclase 9
|
|
chr9_+_45178521
|
14.188
|
|
Fxyd6
|
FXYD domain-containing ion transport regulator 6
|
|
chr8_+_128519001
|
14.186
|
NM_008430
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr11_+_101107726
|
14.165
|
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2
|
|
chr14_-_68742863
|
13.928
|
|
Nefm
|
neurofilament, medium polypeptide
|
|
chr11_+_101107635
|
13.731
|
NM_019444
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2
|
|
chr16_+_91270319
|
13.715
|
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr17_+_24641034
|
13.593
|
|
Caskin1
|
CASK interacting protein 1
|
|
chr13_+_58909168
|
13.164
|
NM_001025074
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr16_+_91270009
|
13.069
|
NM_016968
|
Olig1
|
oligodendrocyte transcription factor 1
|
|
chr12_-_46175433
|
13.064
|
NM_144552
|
Stxbp6
|
syntaxin binding protein 6 (amisyn)
|
|
chr2_+_61884596
|
12.826
|
NM_033552
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10
|
|
chr2_-_115890247
|
12.764
|
NM_001159569
NM_001159570
|
Meis2
|
Meis homeobox 2
|
|
chr8_-_73011217
|
12.708
|
NM_182991
|
Tmem59l
|
transmembrane protein 59-like
|
|
chr16_-_28753298
|
12.691
|
|
Fgf12
|
fibroblast growth factor 12
|
|
chr9_-_97933533
|
12.539
|
NM_022319
|
Clstn2
|
calsyntenin 2
|
|
chr7_+_80885223
|
12.482
|
|
Fam174b
|
family with sequence similarity 174, member B
|
|
chrX_+_20281028
|
12.360
|
NM_145628
|
Usp11
|
ubiquitin specific peptidase 11
|
|
chrX_+_155897136
|
12.310
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr12_+_109572514
|
12.309
|
NM_010010
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1
|
|
chr14_-_124026357
|
12.204
|
NM_177393
|
Nalcn
|
sodium leak channel, non-selective
|
|
chr10_+_62849310
|
12.166
|
NM_013888
|
Dnajc12
|
DnaJ (Hsp40) homolog, subfamily C, member 12
|
|
chr13_+_58909632
|
12.098
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr1_+_164569335
|
12.083
|
|
Myoc
|
myocilin
|
|
chr16_-_45724720
|
11.969
|
|
Tagln3
|
transgelin 3
|
|
chr6_-_136824217
|
11.703
|
|
Mgp
|
matrix Gla protein
|
|
chr1_-_82287990
|
11.640
|
NM_010570
|
Irs1
|
insulin receptor substrate 1
|
|
chr7_-_134164627
|
11.604
|
NM_001102563
|
Prrt2
|
proline-rich transmembrane protein 2
|
|
chrX_+_71550562
|
11.590
|
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1
|
|
chr5_-_77837037
|
11.532
|
NM_001159518
NM_008048
|
Igfbp7
|
insulin-like growth factor binding protein 7
|
|
chr11_-_4847968
|
11.528
|
NM_010904
|
Nefh
|
neurofilament, heavy polypeptide
|
|
chr12_+_16660521
|
11.518
|
|
Ntsr2
|
neurotensin receptor 2
|
|
chr15_-_37389073
|
11.370
|
NM_001170867
NM_001170868
|
Ncald
|
neurocalcin delta
|
|
chr8_+_96676506
|
11.325
|
NM_013603
|
Mt3
|
metallothionein 3
|
|
chr8_-_87461251
|
11.316
|
NM_019945
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr16_-_45724422
|
11.270
|
NM_019754
|
Tagln3
|
transgelin 3
|
|
chr5_-_77836974
|
11.193
|
|
Igfbp7
|
insulin-like growth factor binding protein 7
|
|
chr6_-_113991945
|
11.125
|
NM_009723
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2
|
|
chr6_+_118016385
|
11.006
|
NM_027526
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr14_-_55531726
|
10.900
|
NM_021551
|
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17
|
|
chr3_-_57651409
|
10.808
|
|
Pfn2
|
profilin 2
|
|
chr12_+_16660245
|
10.777
|
NM_008747
|
Ntsr2
|
neurotensin receptor 2
|
|
chr6_-_77929575
|
10.773
|
|
Ctnna2
|
catenin (cadherin associated protein), alpha 2
|
|
chr16_+_41532938
|
10.763
|
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr10_+_115738429
|
10.653
|
NM_029928
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr15_-_63891810
|
10.405
|
NM_144846
|
Fam49b
|
family with sequence similarity 49, member B
|
|
chr1_+_184695026
|
10.343
|
NM_144796
|
Susd4
|
sushi domain containing 4
|
|
chrX_+_155897056
|
10.337
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr14_-_80061976
|
10.156
|
NM_010701
|
Lect1
|
leukocyte cell derived chemotaxin 1
|
|
chr3_-_57651621
|
9.983
|
NM_019410
|
Pfn2
|
profilin 2
|
|
chr4_+_42929523
|
9.973
|
|
N28178
|
expressed sequence N28178
|
|
chr6_+_88775664
|
9.940
|
|
Mgll
|
monoglyceride lipase
|
|
chr7_+_80885175
|
9.861
|
NM_001162532
|
Fam174b
|
family with sequence similarity 174, member B
|
|
chr6_+_83087016
|
9.770
|
NM_009106
NM_133641
|
Rtkn
|
rhotekin
|
|
chr17_-_25036364
|
9.756
|
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3
|
|
chr8_-_113783097
|
9.729
|
NM_009149
|
Glg1
|
golgi apparatus protein 1
|
|
chr2_-_156817751
|
9.716
|
NM_013865
NM_180956
|
Ndrg3
|
N-myc downstream regulated gene 3
|
|
chr3_+_90346956
|
9.594
|
|
S100a16
|
S100 calcium binding protein A16
|
|
chr7_+_25266986
|
9.575
|
NM_153112
|
Cadm4
|
cell adhesion molecule 4
|
|
chr12_+_117724182
|
9.520
|
NM_011215
|
Ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr7_-_31912022
|
9.499
|
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr6_-_92484838
|
9.422
|
NM_001134459
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr2_-_84614960
|
9.411
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr7_-_134164487
|
9.342
|
|
Prrt2
|
proline-rich transmembrane protein 2
|
|
chr8_+_46744023
|
9.315
|
|
Sorbs2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_133054355
|
9.310
|
NM_001081156
|
Trnp1
|
TMF1-regulated nuclear protein 1
|
|
chr19_+_4855178
|
9.287
|
|
Ctsf
|
cathepsin F
|
|
chr18_+_35020867
|
9.277
|
|
Egr1
|
early growth response 1
|
|
chr7_+_134094045
|
9.226
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr10_-_42893894
|
9.194
|
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr6_-_82889694
|
9.130
|
NM_001113481
NM_011350
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain
|
|
chr6_+_107479719
|
9.112
|
NM_008516
|
Lrrn1
|
leucine rich repeat protein 1, neuronal
|
|
chr8_-_75877407
|
9.112
|
|
Large
|
like-glycosyltransferase
|
|
chr7_+_3303657
|
8.990
|
NM_011102
|
Prkcc
|
protein kinase C, gamma
|
|
chr13_-_69878721
|
8.883
|
NM_001145162
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chr6_-_23789259
|
8.882
|
NM_153163
|
Cadps2
|
Ca2+-dependent activator protein for secretion 2
|
|
chr1_+_11404185
|
8.847
|
NM_001160369
NM_001160370
NM_001160371
NM_177173
|
A830018L16Rik
|
RIKEN cDNA A830018L16 gene
|
|
chr19_-_38199291
|
8.827
|
NM_011255
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr1_+_66221851
|
8.826
|
NM_001039934
NM_008632
|
Mtap2
|
microtubule-associated protein 2
|
|
chr1_+_160292665
|
8.796
|
|
Astn1
|
astrotactin 1
|
|
chr1_-_166388431
|
8.766
|
NM_009721
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr8_-_87461220
|
8.760
|
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr16_+_41533317
|
8.724
|
NM_175548
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr3_+_156224757
|
8.720
|
NM_001039094
NM_177274
|
Negr1
|
neuronal growth regulator 1
|
|
chr1_-_166387998
|
8.689
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr1_+_50984361
|
8.628
|
NM_019790
|
Tmeff2
|
transmembrane protein with EGF-like and two follistatin-like domains 2
|
|
chrX_+_50911075
|
8.624
|
NM_028375
|
Cxx1c
|
CAAX box 1 homolog C (human)
|
|
chrX_+_133200913
|
8.562
|
NM_001029978
|
Tceal3
|
transcription elongation factor A (SII)-like 3
|
|
chr7_-_31911884
|
8.549
|
NM_011322
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr1_+_72871060
|
8.441
|
NM_008342
|
Igfbp2
|
insulin-like growth factor binding protein 2
|
|
chr15_-_91403685
|
8.421
|
NM_001033633
|
Slc2a13
|
solute carrier family 2 (facilitated glucose transporter), member 13
|
|
chr2_-_71384748
|
8.404
|
NM_010054
|
Dlx2
|
distal-less homeobox 2
|
|
chr2_+_119625463
|
8.376
|
|
Tyro3
|
TYRO3 protein tyrosine kinase 3
|
|
chr19_+_6427922
|
8.316
|
NM_020253
|
Nrxn2
|
neurexin II
|
|
chr15_+_34167780
|
8.197
|
NM_033521
|
Laptm4b
|
lysosomal-associated protein transmembrane 4B
|
|
chr14_-_80061875
|
8.176
|
|
Lect1
|
leukocyte cell derived chemotaxin 1
|
|
chr15_+_38908403
|
8.174
|
NM_026778
|
Cthrc1
|
collagen triple helix repeat containing 1
|
|
chr7_+_129432810
|
8.124
|
|
Prkcb
|
protein kinase C, beta
|
|
chr7_-_31911754
|
8.036
|
|
Scn1b
|
sodium channel, voltage-gated, type I, beta
|
|
chr16_-_4419805
|
8.019
|
|
Adcy9
|
adenylate cyclase 9
|
|
chr10_-_42894259
|
8.003
|
NM_175407
|
Sobp
|
sine oculis-binding protein homolog (Drosophila)
|
|
chr18_-_16967100
|
8.002
|
|
Cdh2
|
cadherin 2
|
|
chr7_+_129432585
|
7.987
|
NM_008855
|
Prkcb
|
protein kinase C, beta
|
|
chr6_+_117961386
|
7.970
|
|
Rasgef1a
|
RasGEF domain family, member 1A
|
|
chr1_-_175297624
|
7.959
|
|
Cadm3
|
cell adhesion molecule 3
|
|
chr18_-_43059156
|
7.909
|
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr1_-_175297778
|
7.907
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chrX_-_50973482
|
7.860
|
NM_001018063
|
Cxx1b
|
CAAX box 1 homolog B (human)
|
|
chrX_-_100381703
|
7.709
|
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr9_-_97933599
|
7.704
|
|
Clstn2
|
calsyntenin 2
|
|
chr4_+_129662284
|
7.702
|
NM_173071
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr13_-_12199109
|
7.644
|
NM_023868
|
Ryr2
|
ryanodine receptor 2, cardiac
|
|
chr4_-_147412872
|
7.617
|
NM_011929
|
Clcn6
|
chloride channel 6
|
|
chr14_+_66587319
|
7.614
|
NM_013492
|
Clu
|
clusterin
|
|
chr16_-_20621287
|
7.593
|
NM_028420
|
Camk2n2
|
calcium/calmodulin-dependent protein kinase II inhibitor 2
|
|
chr4_+_129662110
|
7.539
|
|
Bai2
|
brain-specific angiogenesis inhibitor 2
|
|
chr6_-_82889660
|
7.393
|
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain
|
|
chr19_-_38199036
|
7.378
|
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr9_-_52487410
|
7.356
|
NM_178906
|
AI593442
|
expressed sequence AI593442
|
|
chrX_+_71550301
|
7.355
|
NM_010273
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1
|
|
chr2_-_23904343
|
7.325
|
|
Hnmt
|
histamine N-methyltransferase
|
|
chr16_+_17276720
|
7.323
|
|
Tmem191c
|
transmembrane protein 191C
|
|
chr9_-_58884114
|
7.281
|
NM_001042752
NM_008684
|
Neo1
|
neogenin
|
|
chr2_-_5635517
|
7.266
|
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr19_-_40525876
|
7.191
|
NM_001034962
NM_001034963
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr7_-_31836457
|
7.187
|
NM_022007
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7
|
|
chrX_+_71550669
|
7.169
|
|
Gdi1
|
guanosine diphosphate (GDP) dissociation inhibitor 1
|
|
chr13_-_78338131
|
7.160
|
NM_010151
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr18_+_57292743
|
7.146
|
NM_001001979
|
Megf10
|
multiple EGF-like-domains 10
|
|
chr19_-_38199267
|
7.094
|
|
Rbp4
|
retinol binding protein 4, plasma
|
|
chr17_-_27650285
|
7.076
|
|
Grm4
|
glutamate receptor, metabotropic 4
|
|
chr10_+_81038028
|
7.063
|
NM_019725
|
Tle2
|
transducin-like enhancer of split 2, homolog of Drosophila E(spl)
|
|
chr6_+_125095236
|
7.057
|
NM_178787
NM_001039669
|
Iffo1
|
intermediate filament family orphan 1
|
|
chr4_-_147412919
|
7.045
|
|
Clcn6
|
chloride channel 6
|
|
chr9_+_110147867
|
7.029
|
|
Cspg5
|
chondroitin sulfate proteoglycan 5
|
|
chr15_+_39491073
|
6.929
|
|
Rims2
|
regulating synaptic membrane exocytosis 2
|
|
chr9_+_75162059
|
6.897
|
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr15_+_54402988
|
6.892
|
|
Mal2
|
mal, T-cell differentiation protein 2
|
|
chr2_-_113669243
|
6.836
|
NM_009162
|
Scg5
|
secretogranin V
|
|
chr1_-_84692750
|
6.801
|
NM_152915
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chr8_-_107995252
|
6.766
|
|
Tppp3
|
tubulin polymerization-promoting protein family member 3
|
|
chr7_-_74904546
|
6.726
|
NM_183312
NM_201639
NM_207663
|
Synm
|
synemin, intermediate filament protein
|
|
chr2_+_119625249
|
6.704
|
NM_019392
|
Tyro3
|
TYRO3 protein tyrosine kinase 3
|
|
chr18_+_37974732
|
6.688
|
NM_033582
|
Pcdhgc4
|
protocadherin gamma subfamily C, 4
|
|
chrX_-_133403050
|
6.670
|
NM_176971
|
Rab9b
|
RAB9B, member RAS oncogene family
|
|
chr2_-_113669229
|
6.645
|
|
Scg5
|
secretogranin V
|
|
chr2_-_12932442
|
6.644
|
NM_153155
|
C1ql3
|
C1q-like 3
|
|
chr11_+_3414884
|
6.635
|
|
Selm
|
selenoprotein M
|
|
chr12_+_70298163
|
6.613
|
|
9330151L19Rik
|
RIKEN cDNA 9330151L19 gene
|
|
chr14_+_34736806
|
6.600
|
|
Gdf10
|
growth differentiation factor 10
|
|
chr7_+_106415956
|
6.548
|
NM_001048167
NM_010837
|
Mtap6
|
microtubule-associated protein 6
|
|
chr11_+_3414792
|
6.543
|
|
Selm
|
selenoprotein M
|
|
chr5_-_133019167
|
6.528
|
|
Auts2
|
autism susceptibility candidate 2
|