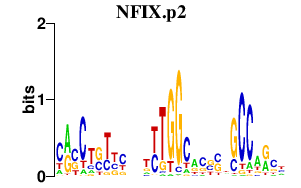
|
chr1_+_154802144
|
19.469
|
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr1_+_154802198
|
18.381
|
NM_175460
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr12_+_30687597
|
18.188
|
|
Pxdn
|
peroxidasin homolog (Drosophila)
|
|
chr1_+_154802091
|
16.254
|
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2
|
|
chr7_+_138079669
|
13.731
|
NM_019564
|
Htra1
|
HtrA serine peptidase 1
|
|
chr10_-_79602040
|
12.757
|
|
Dos
|
downstream of Stk11
|
|
chr18_-_33623400
|
11.616
|
NM_001109989
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr13_-_34222209
|
10.945
|
NM_023716
|
Tubb2b
|
tubulin, beta 2B
|
|
chr18_-_33623287
|
10.551
|
NM_053078
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr18_-_33623668
|
10.386
|
NM_001109988
|
D0H4S114
|
DNA segment, human D4S114
|
|
chr1_+_94728217
|
10.371
|
NM_016696
|
Gpc1
|
glypican 1
|
|
chr6_-_124807733
|
9.993
|
|
Leprel2
|
leprecan-like 2
|
|
chr17_-_23814399
|
9.964
|
|
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a
|
|
chr1_+_94728419
|
9.891
|
|
Gpc1
|
glypican 1
|
|
chr17_-_23814415
|
9.757
|
NM_001161746
NM_013749
|
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a
|
|
chr10_+_24315247
|
8.596
|
NM_010217
|
Ctgf
|
connective tissue growth factor
|
|
chr16_-_85173978
|
8.528
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr16_-_85173851
|
8.449
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr7_-_52347489
|
8.444
|
NM_026555
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr7_-_106501642
|
8.212
|
NM_001111043
NM_001111044
NM_009825
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr13_-_69878721
|
7.938
|
NM_001145162
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1
|
|
chrX_+_132528317
|
7.871
|
NM_029823
|
6530401D17Rik
2900062L11Rik
|
RIKEN cDNA 6530401D17 gene
RIKEN cDNA 2900062L11 gene
|
|
chr7_-_52347012
|
7.706
|
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain
|
|
chr11_-_4060222
|
7.690
|
NM_001164620
NM_177616
|
Ccdc157
|
coiled-coil domain containing 157
|
|
chrX_+_132567905
|
7.235
|
NM_029541
|
6530401D17Rik
|
RIKEN cDNA 6530401D17 gene
|
|
chr11_+_61318701
|
7.176
|
|
B9d1
|
B9 protein domain 1
|
|
chr16_-_85173944
|
7.079
|
NM_001198823
NM_001198824
NM_001198825
NM_001198826
NM_007471
|
App
|
amyloid beta (A4) precursor protein
|
|
chr13_-_78338131
|
6.757
|
NM_010151
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1
|
|
chr18_-_16966954
|
6.740
|
|
Cdh2
|
cadherin 2
|
|
chr12_+_110783786
|
6.736
|
|
Meg3
|
maternally expressed 3
|
|
chr2_-_151458256
|
6.684
|
NM_198214
|
Snph
|
syntaphilin
|
|
chr10_+_28863801
|
6.656
|
NM_026138
|
6330407J23Rik
|
RIKEN cDNA 6330407J23 gene
|
|
chr15_+_32850477
|
6.639
|
NM_008304
|
Sdc2
|
syndecan 2
|
|
chr7_-_106501620
|
6.498
|
|
Serpinh1
|
serine (or cysteine) peptidase inhibitor, clade H, member 1
|
|
chr16_-_85173930
|
6.356
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr1_+_176431933
|
6.346
|
NM_019445
|
Fmn2
|
formin 2
|
|
chr13_-_69138418
|
6.327
|
NM_153534
|
Adcy2
|
adenylate cyclase 2
|
|
chr9_-_99336923
|
6.318
|
NM_008624
|
Mras
|
muscle and microspikes RAS
|
|
chr11_+_61318645
|
6.271
|
NM_013717
|
B9d1
|
B9 protein domain 1
|
|
chr10_-_80945445
|
6.198
|
NM_176954
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr1_-_63223190
|
6.017
|
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1
|
|
chr16_-_85173918
|
5.968
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr7_+_27092187
|
5.947
|
NM_009997
|
Cyp2a4
|
cytochrome P450, family 2, subfamily a, polypeptide 4
|
|
chr2_+_32455168
|
5.941
|
NM_001025311
|
St6galnac6
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 6
|
|
chr1_-_63223345
|
5.773
|
NM_001160038
NM_001160039
NM_001160040
NM_145518
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1
|
|
chr16_+_11066390
|
5.767
|
NM_009223
|
Snn
|
stannin
|
|
chr17_-_14831095
|
5.689
|
|
Thbs2
|
thrombospondin 2
|
|
chr1_-_63223287
|
5.621
|
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1
|
|
chr5_-_31456576
|
5.554
|
NM_008622
|
Mpv17
|
MpV17 mitochondrial inner membrane protein
|
|
chr19_+_10057131
|
5.508
|
NM_010239
|
Fth1
|
ferritin heavy chain 1
|
|
chr18_+_37821598
|
5.486
|
NM_033584
|
Pcdhga1
|
protocadherin gamma subfamily A, 1
|
|
chr12_+_110806715
|
5.424
|
|
Meg3
|
maternally expressed 3
|
|
chr12_+_110783593
|
5.316
|
|
Meg3
|
maternally expressed 3
|
|
chr7_+_27620354
|
5.243
|
NM_007812
|
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr10_-_80945379
|
5.238
|
|
Celf5
|
CUGBP, Elav-like family member 5
|
|
chr8_-_97666364
|
5.193
|
NM_001145831
NM_010631
|
Kifc3
|
kinesin family member C3
|
|
chr6_-_55124983
|
5.157
|
NM_009349
|
Inmt
|
indolethylamine N-methyltransferase
|
|
chr17_-_25974021
|
5.118
|
NM_144816
|
Rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila)
|
|
chr2_+_35547418
|
4.831
|
NM_001001602
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr6_+_34426324
|
4.798
|
|
Bpgm
|
2,3-bisphosphoglycerate mutase
|
|
chr11_+_104092809
|
4.788
|
|
Mapt
|
microtubule-associated protein tau
|
|
chr6_+_34426288
|
4.719
|
|
Bpgm
|
2,3-bisphosphoglycerate mutase
|
|
chr9_-_58388489
|
4.694
|
NM_133983
|
Cd276
|
CD276 antigen
|
|
chr2_+_106533281
|
4.681
|
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr1_-_25886551
|
4.533
|
NM_175642
|
Bai3
|
brain-specific angiogenesis inhibitor 3
|
|
chr7_+_27620380
|
4.503
|
|
Cyp2a4
Cyp2a5
|
cytochrome P450, family 2, subfamily a, polypeptide 4
cytochrome P450, family 2, subfamily a, polypeptide 5
|
|
chr7_+_29052821
|
4.469
|
NM_198425
|
Eid2
|
EP300 interacting inhibitor of differentiation 2
|
|
chr9_-_121404799
|
4.369
|
NM_031161
|
Cck
|
cholecystokinin
|
|
chr13_+_13529868
|
4.293
|
NM_010917
|
Nid1
|
nidogen 1
|
|
chr6_+_34426352
|
4.282
|
NM_007563
|
Bpgm
|
2,3-bisphosphoglycerate mutase
|
|
chr2_+_156301651
|
4.165
|
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr15_-_93426312
|
4.068
|
NM_001033217
|
Prickle1
|
prickle homolog 1 (Drosophila)
|
|
chr1_-_174136874
|
4.058
|
NM_011063
|
Pea15a
|
phosphoprotein enriched in astrocytes 15A
|
|
chr12_-_71448600
|
4.058
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr13_+_13529902
|
3.971
|
|
Nid1
|
nidogen 1
|
|
chr5_-_92471976
|
3.964
|
NM_177270
NM_016912
|
Cdkl2
|
cyclin-dependent kinase-like 2 (CDC2-related kinase)
|
|
chr9_+_107838164
|
3.890
|
NM_145621
|
Camkv
|
CaM kinase-like vesicle-associated
|
|
chr13_+_13529923
|
3.808
|
|
Nid1
|
nidogen 1
|
|
chr19_+_21852816
|
3.781
|
NM_001033759
NM_031997
|
Tmem2
|
transmembrane protein 2
|
|
chr2_+_106533615
|
3.767
|
NM_001143683
|
Mpped2
|
metallophosphoesterase domain containing 2
|
|
chr10_+_81022283
|
3.756
|
|
Aes
|
amino-terminal enhancer of split
|
|
chr11_+_3414983
|
3.749
|
|
Selm
|
selenoprotein M
|
|
chr11_+_3414792
|
3.746
|
|
Selm
|
selenoprotein M
|
|
chr16_-_22439642
|
3.745
|
NM_023794
|
Etv5
|
ets variant gene 5
|
|
chr14_+_8835385
|
3.680
|
NM_025341
|
Abhd6
|
abhydrolase domain containing 6
|
|
chr3_+_88425073
|
3.677
|
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr4_-_24778203
|
3.625
|
NM_001033531
NM_001163020
|
Klhl32
|
kelch-like 32 (Drosophila)
|
|
chr10_+_77441365
|
3.586
|
NM_026431
|
1810043G02Rik
|
RIKEN cDNA 1810043G02 gene
|
|
chr5_-_134718500
|
3.572
|
|
Gtf2i
|
general transcription factor II I
|
|
chr3_+_88425352
|
3.564
|
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr5_+_117807290
|
3.548
|
NM_021539
|
Wsb2
|
WD repeat and SOCS box-containing 2
|
|
chr2_+_156301543
|
3.531
|
NM_001006664
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1
|
|
chr3_+_88425294
|
3.488
|
NM_001198912
NM_001198913
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr7_-_31836457
|
3.459
|
NM_022007
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7
|
|
chr4_-_115320309
|
3.448
|
NM_007823
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1
|
|
chr3_+_88425011
|
3.409
|
NM_008487
|
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2
|
|
chr15_-_32174172
|
3.337
|
|
Snord123
|
small nucleolar RNA, C/D box 123
|
|
chr10_+_110357230
|
3.333
|
NM_007792
|
Csrp2
|
cysteine and glycine-rich protein 2
|
|
chr7_-_97605681
|
3.253
|
NM_025460
|
Tmem126a
|
transmembrane protein 126A
|
|
chrX_+_132804834
|
3.248
|
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr4_-_45543623
|
3.245
|
NM_001033306
|
Shb
|
src homology 2 domain-containing transforming protein B
|
|
chr10_+_81022304
|
3.236
|
NM_010347
|
Aes
|
amino-terminal enhancer of split
|
|
chr11_+_3414686
|
3.217
|
NM_053267
|
Selm
|
selenoprotein M
|
|
chr10_+_81022229
|
3.192
|
|
Aes
|
amino-terminal enhancer of split
|
|
chr11_+_104092793
|
3.115
|
|
Mapt
|
microtubule-associated protein tau
|
|
chr11_+_45869506
|
3.085
|
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr6_+_54631644
|
2.954
|
NM_026629
|
2410066E13Rik
|
RIKEN cDNA 2410066E13 gene
|
|
chr14_-_21871345
|
2.906
|
|
Ap3m1
|
adaptor-related protein complex 3, mu 1 subunit
|
|
chr1_-_79855209
|
2.882
|
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr16_-_10314010
|
2.855
|
NM_007929
|
Emp2
|
epithelial membrane protein 2
|
|
chr11_+_3414884
|
2.817
|
|
Selm
|
selenoprotein M
|
|
chr2_+_32502186
|
2.793
|
|
Eng
|
endoglin
|
|
chr8_-_41596638
|
2.784
|
NM_011135
|
Cnot7
|
CCR4-NOT transcription complex, subunit 7
|
|
chr6_-_125444722
|
2.764
|
NM_007657
|
Cd9
|
CD9 antigen
|
|
chr7_+_80885175
|
2.709
|
NM_001162532
|
Fam174b
|
family with sequence similarity 174, member B
|
|
chr18_+_37911236
|
2.696
|
NM_033579
|
Pcdhgb7
|
protocadherin gamma subfamily B, 7
|
|
chr6_+_48791616
|
2.682
|
NM_001098271
|
Tmem176a
|
transmembrane protein 176A
|
|
chr6_-_125444695
|
2.666
|
|
Cd9
|
CD9 antigen
|
|
chr15_-_101616888
|
2.641
|
NM_213728
|
Krt72-ps
|
keratin 72, pseudogene
|
|
chr11_-_116007231
|
2.610
|
|
Fbf1
|
Fas (TNFRSF6) binding factor 1
|
|
chr6_+_117791285
|
2.591
|
|
Zfp637
|
zinc finger protein 637
|
|
chr8_+_97197870
|
2.558
|
|
Arl2bp
|
ADP-ribosylation factor-like 2 binding protein
|
|
chr1_-_175297624
|
2.536
|
|
Cadm3
|
cell adhesion molecule 3
|
|
chr8_-_34752419
|
2.505
|
NM_178648
|
Ubxn8
|
UBX domain protein 8
|
|
chr1_-_173080710
|
2.495
|
NM_025321
|
Sdhc
|
succinate dehydrogenase complex, subunit C, integral membrane protein
|
|
chr8_+_87162885
|
2.492
|
|
Cacna1a
|
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit
|
|
chr5_-_114250360
|
2.486
|
NM_146162
|
Tmem119
|
transmembrane protein 119
|
|
chr1_-_175297778
|
2.442
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chr12_+_70994091
|
2.430
|
NM_178628
|
Atl1
|
atlastin GTPase 1
|
|
chr6_-_92517359
|
2.407
|
NM_001134461
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr19_-_58529766
|
2.404
|
|
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1
|
|
chr18_+_37901716
|
2.397
|
NM_033578
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6
|
|
chr18_+_38115192
|
2.395
|
NM_153793
|
Rell2
|
RELT-like 2
|
|
chrX_-_132749976
|
2.389
|
NM_009052
|
Bex1
|
brain expressed gene 1
|
|
chr16_-_85173764
|
2.379
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr1_-_79855229
|
2.344
|
NM_009255
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2
|
|
chr7_-_148008029
|
2.334
|
NM_001001180
|
Zfp941
|
zinc finger protein 941
|
|
chr17_-_14831058
|
2.330
|
|
Thbs2
|
thrombospondin 2
|
|
chr9_+_56266415
|
2.313
|
|
Hmg20a
|
high mobility group 20A
|
|
chr1_+_66221851
|
2.307
|
NM_001039934
NM_008632
|
Mtap2
|
microtubule-associated protein 2
|
|
chr5_-_4104331
|
2.233
|
|
Cyp51
|
cytochrome P450, family 51
|
|
chr11_+_104092749
|
2.226
|
NM_001038609
NM_010838
|
Mapt
|
microtubule-associated protein tau
|
|
chr2_+_70400179
|
2.225
|
NM_008077
|
Gad1
|
glutamic acid decarboxylase 1
|
|
chr1_+_59821480
|
2.201
|
NM_007561
|
Bmpr2
|
bone morphogenic protein receptor, type II (serine/threonine kinase)
|
|
chr6_-_124807704
|
2.190
|
NM_013534
|
Leprel2
|
leprecan-like 2
|
|
chr16_-_10313963
|
2.174
|
|
Emp2
|
epithelial membrane protein 2
|
|
chr10_-_105011265
|
2.169
|
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chr13_-_69138229
|
2.160
|
|
Adcy2
|
adenylate cyclase 2
|
|
chr10_+_75398588
|
2.154
|
|
Chchd10
|
coiled-coil-helix-coiled-coil-helix domain containing 10
|
|
chr3_-_57379831
|
2.147
|
NM_001168281
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr15_-_76881097
|
2.142
|
NM_013593
|
Mb
|
myoglobin
|
|
chr3_-_152003317
|
2.131
|
NM_001162375
NM_174868
|
Fam73a
|
family with sequence similarity 73, member A
|
|
chrX_+_132804791
|
2.118
|
NM_001110233
NM_001110234
NM_009750
|
Ngfrap1
|
nerve growth factor receptor (TNFRSF16) associated protein 1
|
|
chr13_-_34437042
|
2.117
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr3_+_108394195
|
2.085
|
NM_181400
|
Wdr47
|
WD repeat domain 47
|
|
chr1_-_94370312
|
2.072
|
NM_024197
|
Ndufa10
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10
|
|
chr10_+_79174337
|
2.069
|
|
Bsg
|
basigin
|
|
chr3_-_57379600
|
2.061
|
NM_133784
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr18_+_37890662
|
2.035
|
NM_033577
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5
|
|
chr12_+_76409299
|
2.027
|
NM_023275
|
Rhoj
|
ras homolog gene family, member J
|
|
chr7_+_150659625
|
2.015
|
NM_008767
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18
|
|
chr1_+_168184269
|
2.007
|
NM_001164528
|
Ildr2
|
immunoglobulin-like domain containing receptor 2
|
|
chr5_-_143026961
|
1.971
|
NM_178702
|
Radil
|
Ras association and DIL domains
|
|
chr15_+_58247016
|
1.965
|
NM_145959
|
D15Ertd621e
|
DNA segment, Chr 15, ERATO Doi 621, expressed
|
|
chr10_-_105011439
|
1.959
|
NM_177368
|
Tmtc2
|
transmembrane and tetratricopeptide repeat containing 2
|
|
chrX_-_147531030
|
1.958
|
NM_001164578
NM_175146
|
Tsr2
|
TSR2, 20S rRNA accumulation, homolog (S. cerevisiae)
|
|
chr8_-_93659394
|
1.950
|
NM_010241
|
Aktip
|
thymoma viral proto-oncogene 1 interacting protein
|
|
chr5_-_114541313
|
1.947
|
NM_026805
|
Svop
|
SV2 related protein
|
|
chr8_-_28239121
|
1.934
|
|
Brf2
|
BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like
|
|
chr9_+_122714664
|
1.929
|
NM_001013405
|
D9Ertd402e
|
DNA segment, Chr 9, ERATO Doi 402, expressed
|
|
chr7_-_31230577
|
1.910
|
NM_007467
|
Aplp1
|
amyloid beta (A4) precursor-like protein 1
|
|
chr5_+_118426774
|
1.901
|
NM_145564
|
Fbxo21
|
F-box protein 21
|
|
chr12_-_80273320
|
1.889
|
NM_016800
|
Vti1b
|
vesicle transport through interaction with t-SNAREs 1B homolog
|
|
chr16_-_30550622
|
1.885
|
NM_172614
|
Tmem44
|
transmembrane protein 44
|
|
chr13_-_69138164
|
1.830
|
|
Adcy2
|
adenylate cyclase 2
|
|
chr19_-_10630215
|
1.829
|
|
Tmem216
|
transmembrane protein 216
|
|
chr11_-_20731016
|
1.826
|
NM_173752
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr2_-_151458299
|
1.804
|
|
Snph
|
syntaphilin
|
|
chr1_+_74589461
|
1.783
|
NM_001081456
|
Plcd4
|
phospholipase C, delta 4
|
|
chr8_-_28239077
|
1.777
|
NM_025686
|
Brf2
|
BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like
|
|
chr13_+_13529947
|
1.770
|
|
Nid1
|
nidogen 1
|
|
chr8_-_128093602
|
1.760
|
|
Sipa1l2
|
signal-induced proliferation-associated 1 like 2
|
|
chr7_-_137663020
|
1.752
|
NM_001136054
NM_013799
|
Ate1
|
arginyltransferase 1
|
|
chr19_+_45635070
|
1.735
|
NM_172639
|
Dpcd
|
deleted in primary ciliary dyskinesia
|
|
chr5_+_118426721
|
1.733
|
|
Fbxo21
|
F-box protein 21
|
|
chrX_+_8469478
|
1.731
|
NM_175327
|
B630019K06Rik
|
RIKEN cDNA B630019K06 gene
|
|
chr6_+_117791197
|
1.693
|
NM_177684
|
Zfp637
|
zinc finger protein 637
|
|
chr8_-_83404377
|
1.692
|
NM_021356
|
Gab1
|
growth factor receptor bound protein 2-associated protein 1
|
|
chr18_+_38578583
|
1.679
|
NM_022996
|
Ndfip1
|
Nedd4 family interacting protein 1
|
|
chr6_-_7643156
|
1.662
|
NM_012055
|
Asns
|
asparagine synthetase
|
|
chr11_+_45869473
|
1.659
|
NM_009616
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr14_+_121434765
|
1.641
|
NM_134082
|
Farp1
|
FERM, RhoGEF (Arhgef) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_+_126378563
|
1.640
|
NM_011978
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2
|
|
chr7_-_147340532
|
1.633
|
NM_183147
|
Sprn
|
shadow of prion protein
|
|
chr11_+_94871181
|
1.612
|
NM_146025
|
Samd14
|
sterile alpha motif domain containing 14
|
|
chr13_-_25361773
|
1.605
|
NM_009513
|
Nrsn1
|
neurensin 1
|
|
chr3_-_8667013
|
1.590
|
NM_010423
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1
|
|
chr19_+_45101318
|
1.581
|
|
Lzts2
|
leucine zipper, putative tumor suppressor 2
|
|
chr9_-_121404750
|
1.565
|
|
Cck
|
cholecystokinin
|
|
chr10_-_60609031
|
1.560
|
|
Sgpl1
|
sphingosine phosphate lyase 1
|