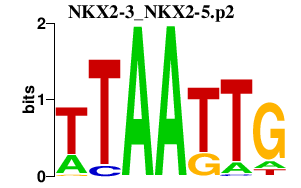
|
chr19_-_44470218
|
11.395
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr15_-_96861590
|
10.931
|
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr11_+_52202137
|
9.128
|
|
Vdac1
|
voltage-dependent anion channel 1
|
|
chr11_+_96212945
|
8.258
|
NM_134032
|
Hoxb2
|
homeobox B2
|
|
chr11_+_75578548
|
8.163
|
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide
|
|
chr2_+_35159780
|
8.138
|
|
Gsn
|
gelsolin
|
|
chr6_+_4484651
|
8.132
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr2_+_125860377
|
7.895
|
NM_008008
|
Fgf7
|
fibroblast growth factor 7
|
|
chr19_-_44476302
|
7.835
|
|
Scd1
|
stearoyl-Coenzyme A desaturase 1
|
|
chr11_+_29607599
|
7.453
|
|
Rtn4
|
reticulon 4
|
|
chr1_+_45368443
|
7.288
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr3_+_54165028
|
7.211
|
NM_001198765
NM_001198766
NM_015784
|
Postn
|
periostin, osteoblast specific factor
|
|
chr2_+_125860507
|
7.055
|
|
Fgf7
|
fibroblast growth factor 7
|
|
chr11_+_67013553
|
6.929
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr11_+_66984528
|
6.799
|
NM_001039545
|
Myh2
|
myosin, heavy polypeptide 2, skeletal muscle, adult
|
|
chr7_-_56103389
|
6.509
|
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr7_-_56103405
|
6.470
|
NM_001198841
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr6_+_83290548
|
6.457
|
|
Mobkl1b
|
MOB1, Mps One Binder kinase activator-like 1B (yeast)
|
|
chr1_-_55135054
|
6.383
|
|
Hspd1
|
heat shock protein 1 (chaperonin)
|
|
chr6_+_4483741
|
6.248
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr15_-_56526052
|
6.161
|
NM_008216
|
Has2
|
hyaluronan synthase 2
|
|
chr1_+_45368382
|
6.026
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr17_-_48571942
|
5.787
|
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2
|
|
chr17_-_48572043
|
5.783
|
NM_009694
|
Apobec2
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 2
|
|
chr10_-_85474581
|
5.695
|
NM_177772
|
Bpil2
|
bactericidal/permeability-increasing protein-like 2
|
|
chr1_-_165243777
|
5.570
|
NM_001025570
NM_011127
NM_175686
|
Prrx1
|
paired related homeobox 1
|
|
chr13_+_49639847
|
5.420
|
|
Aspn
|
asporin
|
|
chrX_+_154137114
|
5.208
|
|
Smpx
|
small muscle protein, X-linked
|
|
chr9_-_103256690
|
5.198
|
|
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3
|
|
chr11_-_119971500
|
5.107
|
|
Slc38a10
|
solute carrier family 38, member 10
|
|
chr8_+_131238027
|
4.838
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr6_+_112223751
|
4.805
|
NM_144799
|
Lmcd1
|
LIM and cysteine-rich domains 1
|
|
chr10_-_116786025
|
4.737
|
|
Cpsf6
|
cleavage and polyadenylation specific factor 6
|
|
chr3_+_67696141
|
4.647
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr19_-_55177562
|
4.587
|
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial
|
|
chr13_+_83643032
|
4.505
|
NM_001170537
NM_025282
|
Mef2c
|
myocyte enhancer factor 2C
|
|
chr9_+_92502076
|
4.474
|
|
|
|
|
chr3_+_159502646
|
4.449
|
NM_026582
|
Wls
|
wntless homolog (Drosophila)
|
|
chr9_+_51855254
|
4.418
|
NM_001104616
|
Rdx
|
radixin
|
|
chr16_+_45094383
|
4.353
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr5_+_104868321
|
4.311
|
|
Spp1
|
secreted phosphoprotein 1
|
|
chr19_+_10271150
|
4.302
|
|
Fads1
|
fatty acid desaturase 1
|
|
chr2_+_74707394
|
4.285
|
|
Mtx2
|
metaxin 2
|
|
chr17_-_14831058
|
4.275
|
|
Thbs2
|
thrombospondin 2
|
|
chrX_+_34337066
|
4.273
|
|
Slc25a5
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 5
|
|
chr6_-_99216494
|
4.017
|
NM_001197321
|
Foxp1
|
forkhead box P1
|
|
chr13_-_102453449
|
3.944
|
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|
|
chr9_-_66880619
|
3.892
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr11_+_29618696
|
3.868
|
|
Rtn4
|
reticulon 4
|
|
chr9_+_108239210
|
3.864
|
|
Rhoa
|
ras homolog gene family, member A
|
|
chr13_+_49639799
|
3.846
|
NM_001172481
NM_025711
|
Aspn
|
asporin
|
|
chrX_-_156560678
|
3.840
|
|
Pdha1
|
pyruvate dehydrogenase E1 alpha 1
|
|
chrX_+_154137049
|
3.766
|
NM_025357
|
Smpx
|
small muscle protein, X-linked
|
|
chr5_-_122907269
|
3.574
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr2_+_132673469
|
3.453
|
NM_025646
|
Crls1
|
cardiolipin synthase 1
|
|
chr17_-_14831095
|
3.448
|
|
Thbs2
|
thrombospondin 2
|
|
chr9_+_54758322
|
3.439
|
|
Ireb2
|
iron responsive element binding protein 2
|
|
chr6_+_54839147
|
3.417
|
|
Znrf2
|
zinc and ring finger 2
|
|
chr2_+_162760505
|
3.407
|
|
|
|
|
chr2_-_84490827
|
3.380
|
NM_001085448
NM_001085449
NM_001085450
NM_007615
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chr4_+_140256449
|
3.347
|
|
Rcc2
|
regulator of chromosome condensation 2
|
|
chr2_+_170336930
|
3.333
|
|
Pfdn4
|
prefoldin 4
|
|
chr6_-_87793898
|
3.316
|
|
Cnbp
|
cellular nucleic acid binding protein
|
|
chr2_+_174171525
|
3.225
|
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus
|
|
chr11_-_75114563
|
3.183
|
|
Rpa1
|
replication protein A1
|
|
chr1_-_165243608
|
3.176
|
|
Prrx1
|
paired related homeobox 1
|
|
chr6_+_4490515
|
3.166
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr3_+_68376289
|
3.161
|
NM_013928
|
Schip1
|
schwannomin interacting protein 1
|
|
chr5_-_89886732
|
3.155
|
NM_008096
|
Gc
|
group specific component
|
|
chr11_+_29618531
|
3.101
|
NM_024226
|
Rtn4
|
reticulon 4
|
|
chr11_-_86331141
|
3.095
|
|
Rps6kb1
|
ribosomal protein S6 kinase, polypeptide 1
|
|
chr10_-_117252126
|
3.087
|
|
|
|
|
chr2_+_34631438
|
3.082
|
|
Hspa5
|
heat shock protein 5
|
|
chr3_+_79690856
|
3.081
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr3_+_68376319
|
2.990
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chr1_+_40082070
|
2.985
|
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr1_+_174051459
|
2.950
|
|
Copa
|
coatomer protein complex subunit alpha
|
|
chr12_+_39510297
|
2.926
|
NM_001163154
|
Etv1
|
ets variant gene 1
|
|
chr1_-_180259336
|
2.903
|
|
Hnrnpu
|
heterogeneous nuclear ribonucleoprotein U
|
|
chr16_-_19706367
|
2.886
|
|
Lamp3
|
lysosomal-associated membrane protein 3
|
|
chr4_+_57907032
|
2.881
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr6_-_52176102
|
2.856
|
|
Hoxa9
|
homeobox A9
|
|
chr8_-_37631772
|
2.850
|
|
Dlc1
|
deleted in liver cancer 1
|
|
chr13_-_74832039
|
2.824
|
|
Cast
|
calpastatin
|
|
chr8_-_71601126
|
2.809
|
|
Slc18a1
|
solute carrier family 18 (vesicular monoamine), member 1
|
|
chr1_+_40082201
|
2.784
|
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr8_-_64381465
|
2.772
|
NM_001081390
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr15_-_81191176
|
2.769
|
NM_011399
|
Slc25a17
|
solute carrier family 25 (mitochondrial carrier, peroxisomal membrane protein), member 17
|
|
chrX_+_132424640
|
2.750
|
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B9
|
|
chr2_-_84490666
|
2.738
|
NM_001085453
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chr11_-_116712559
|
2.706
|
|
Srsf2
|
serine/arginine-rich splicing factor 2
|
|
chr1_+_40496493
|
2.697
|
NM_010743
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr18_+_37834003
|
2.673
|
|
Pcdhga3
|
protocadherin gamma subfamily A, 3
|
|
chr6_+_145882641
|
2.673
|
NM_010656
|
Sspn
|
sarcospan
|
|
chr16_-_11220854
|
2.661
|
|
Gspt1
|
G1 to S phase transition 1
|
|
chr7_+_31548774
|
2.638
|
NM_001166173
NM_001166174
NM_172899
|
Dmkn
|
dermokine
|
|
chr5_+_35341726
|
2.575
|
NM_001163512
|
Rgs12
|
regulator of G-protein signaling 12
|
|
chr18_+_38476622
|
2.573
|
|
|
|
|
chr7_+_134356373
|
2.564
|
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr2_+_13501725
|
2.546
|
|
Vim
|
vimentin
|
|
chr13_+_40955500
|
2.513
|
NM_023887
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme
|
|
chrX_-_35774388
|
2.512
|
|
Lamp2
|
lysosomal-associated membrane protein 2
|
|
chr10_+_87323855
|
2.498
|
NM_001111274
NM_001111276
NM_184052
|
Igf1
|
insulin-like growth factor 1
|
|
chr5_-_137548123
|
2.493
|
NM_008871
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr5_-_114881393
|
2.450
|
|
|
|
|
chr6_-_136890189
|
2.415
|
NM_007486
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta
|
|
chrX_+_130385546
|
2.411
|
NM_022322
|
Tnmd
|
tenomodulin
|
|
chr19_-_47766512
|
2.372
|
NM_007732
|
Col17a1
|
collagen, type XVII, alpha 1
|
|
chr6_-_72383494
|
2.363
|
|
Mat2a
|
methionine adenosyltransferase II, alpha
|
|
chr10_-_88067758
|
2.349
|
NM_175418
|
Mybpc1
|
myosin binding protein C, slow-type
|
|
chr16_-_23520662
|
2.333
|
NM_008555
|
Masp1
|
mannan-binding lectin serine peptidase 1
|
|
chr9_+_92502908
|
2.329
|
|
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2
|
|
chr10_+_56097078
|
2.326
|
NM_010288
|
Gja1
|
gap junction protein, alpha 1
|
|
chr5_-_100391613
|
2.322
|
|
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D
|
|
chr2_+_138104219
|
2.306
|
NM_145534
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr4_+_48572811
|
2.272
|
|
5730528L13Rik
|
RIKEN cDNA 5730528L13 gene
|
|
chr2_+_34631709
|
2.257
|
|
Hspa5
|
heat shock protein 5
|
|
chr9_+_5345475
|
2.255
|
NM_009808
|
Casp12
|
caspase 12
|
|
chr2_+_174171943
|
2.228
|
|
|
|
|
chr15_-_22644738
|
2.227
|
NM_001165971
|
Gm5803
|
predicted gene 5803
|
|
chr16_-_19706392
|
2.225
|
|
Lamp3
|
lysosomal-associated membrane protein 3
|
|
chr5_-_122907002
|
2.219
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr5_-_136384495
|
2.182
|
|
|
|
|
chr13_+_16106279
|
2.168
|
NM_008380
|
Inhba
|
inhibin beta-A
|
|
chr3_+_123008796
|
2.148
|
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae)
|
|
chr5_-_122906470
|
2.136
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr16_+_56477956
|
2.136
|
NM_001014399
NM_001014422
NM_001014423
NM_001014424
NM_178790
|
Abi3bp
|
ABI gene family, member 3 (NESH) binding protein
|
|
chr14_+_102237708
|
2.118
|
|
Lmo7
|
LIM domain only 7
|
|
chr12_+_33535650
|
2.117
|
|
Nampt
|
nicotinamide phosphoribosyltransferase
|
|
chr2_-_26923387
|
2.092
|
NM_001159602
|
Tmem8c
|
transmembrane protein 8C
|
|
chr1_+_40497472
|
2.084
|
NM_001025602
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr2_+_34631124
|
2.077
|
|
Hspa5
|
heat shock protein 5
|
|
chr16_-_19706433
|
2.065
|
NM_177356
|
Lamp3
|
lysosomal-associated membrane protein 3
|
|
chr19_-_5845001
|
2.050
|
|
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr13_-_104554635
|
2.044
|
NM_172592
|
Srsf12
|
serine/arginine-rich splicing factor 12
|
|
chr6_+_55027608
|
1.997
|
|
Gars
|
glycyl-tRNA synthetase
|
|
chr1_-_151808361
|
1.975
|
NM_008869
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent)
|
|
chr8_+_48907496
|
1.932
|
NM_001111318
|
Cldn24
|
claudin 22-like
|
|
chr3_-_57098670
|
1.923
|
|
Tm4sf1
|
transmembrane 4 superfamily member 1
|
|
chr11_-_86399926
|
1.923
|
|
Tmem49
|
transmembrane protein 49
|
|
chr19_+_55700193
|
1.898
|
|
|
|
|
chr4_+_6321713
|
1.888
|
|
Sdcbp
|
syndecan binding protein
|
|
chr19_-_5844533
|
1.877
|
|
Neat1
|
nuclear paraspeckle assembly transcript 1 (non-protein coding)
|
|
chr15_+_78860043
|
1.857
|
|
H1f0
|
H1 histone family, member 0
|
|
chr13_+_55501008
|
1.851
|
NM_011392
|
Slc34a1
|
solute carrier family 34 (sodium phosphate), member 1
|
|
chr4_+_118906433
|
1.849
|
NM_001042411
|
Lepre1
|
leprecan 1
|
|
chr6_+_112409498
|
1.838
|
NM_007617
|
Cav3
|
caveolin 3
|
|
chr11_-_120668581
|
1.825
|
|
Fasn
|
fatty acid synthase
|
|
chr9_-_110649625
|
1.822
|
NM_011199
|
Pth1r
|
parathyroid hormone 1 receptor
|
|
chr1_+_137904795
|
1.799
|
NM_001160145
|
Tmem9
|
transmembrane protein 9
|
|
chr5_+_147760198
|
1.795
|
NM_025652
|
Gtf3a
|
general transcription factor III A
|
|
chr6_+_34659440
|
1.771
|
NM_145575
|
Cald1
|
caldesmon 1
|
|
chr7_-_149762551
|
1.751
|
|
H19
|
H19 fetal liver mRNA
|
|
chr13_-_115248787
|
1.747
|
|
Fst
|
follistatin
|
|
chr2_-_119916315
|
1.740
|
|
Ehd4
|
EH-domain containing 4
|
|
chr4_-_129055102
|
1.715
|
|
Zbtb8a
|
zinc finger and BTB domain containing 8a
|
|
chr1_+_158769044
|
1.704
|
NM_028333
|
Angptl1
|
angiopoietin-like 1
|
|
chr8_-_112103065
|
1.670
|
NM_017370
|
Hp
|
haptoglobin
|
|
chr1_+_51346086
|
1.669
|
|
Sdpr
|
serum deprivation response
|
|
chr5_+_52755183
|
1.642
|
|
Sod3
|
superoxide dismutase 3, extracellular
|
|
chr5_+_88354611
|
1.634
|
NM_007786
|
Csn3
|
casein kappa
|
|
chr2_+_13504271
|
1.628
|
|
|
|
|
chr6_+_112409684
|
1.624
|
|
Cav3
|
caveolin 3
|
|
chr11_+_72264668
|
1.619
|
|
Mybbp1a
|
MYB binding protein (P160) 1a
|
|
chr6_+_115372083
|
1.603
|
NM_011146
|
Pparg
|
peroxisome proliferator activated receptor gamma
|
|
chr9_-_79566271
|
1.594
|
NM_007730
|
Col12a1
|
collagen, type XII, alpha 1
|
|
chr11_+_96160484
|
1.587
|
NM_008269
|
Hoxb6
|
homeobox B6
|
|
chr14_-_73762840
|
1.579
|
|
Itm2b
|
integral membrane protein 2B
|
|
chr11_+_52202706
|
1.574
|
|
Vdac1
|
voltage-dependent anion channel 1
|
|
chr3_+_60432922
|
1.570
|
|
Mbnl1
|
muscleblind-like 1 (Drosophila)
|
|
chr11_+_79845841
|
1.565
|
|
Suz12
|
suppressor of zeste 12 homolog (Drosophila)
|
|
chr19_-_4877874
|
1.562
|
NM_013456
|
Actn3
|
actinin alpha 3
|
|
chr15_-_103045710
|
1.544
|
NM_001110216
NM_007626
|
Cbx5
|
chromobox homolog 5 (Drosophila HP1a)
|
|
chr4_+_57907600
|
1.523
|
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr1_+_109319264
|
1.511
|
NM_027548
|
Serpinb7
|
serine (or cysteine) peptidase inhibitor, clade B, member 7
|
|
chr11_+_96160268
|
1.508
|
|
Hoxb6
|
homeobox B6
|
|
chr1_+_90003410
|
1.505
|
|
Ugt1a6b
|
UDP glucuronosyltransferase 1 family, polypeptide A6B
|
|
chr5_-_136174262
|
1.501
|
|
|
|
|
chr6_-_136889984
|
1.501
|
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta
|
|
chr1_-_172018982
|
1.499
|
NM_022563
|
Ddr2
|
discoidin domain receptor family, member 2
|
|
chr14_-_61877218
|
1.498
|
NM_011892
|
Sgcg
|
sarcoglycan, gamma (dystrophin-associated glycoprotein)
|
|
chr2_+_162758440
|
1.494
|
|
Srsf6
|
serine/arginine-rich splicing factor 6
|
|
chr9_-_78326946
|
1.494
|
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1
|
|
chr5_-_108304062
|
1.490
|
NM_007964
|
Evi5
|
ecotropic viral integration site 5
|
|
chr11_-_74991244
|
1.486
|
|
Ovca2
|
candidate tumor suppressor in ovarian cancer 2
|
|
chrX_+_156278399
|
1.481
|
NM_001135728
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr4_-_132383213
|
1.466
|
|
Ppp1r8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8
|
|
chr5_-_17341534
|
1.465
|
|
Cd36
|
CD36 antigen
|
|
chr18_+_61085283
|
1.449
|
NM_177407
|
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha
|
|
chr2_-_110203264
|
1.449
|
|
Fibin
|
fin bud initiation factor homolog (zebrafish)
|
|
chr2_+_35413977
|
1.439
|
NM_001114125
|
Dab2ip
|
disabled homolog 2 (Drosophila) interacting protein
|
|
chr4_+_8533630
|
1.438
|
|
Rab2a
|
RAB2A, member RAS oncogene family
|
|
chr2_+_109516053
|
1.433
|
NM_001048139
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr15_-_96850752
|
1.432
|
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr17_-_14831233
|
1.427
|
|
Thbs2
|
thrombospondin 2
|
|
chr1_+_136186579
|
1.426
|
NM_031189
|
Myog
|
myogenin
|
|
chr2_+_173603706
|
1.420
|
|
Vapb
|
vesicle-associated membrane protein, associated protein B and C
|
|
chr11_+_51085400
|
1.418
|
|
Clk4
|
CDC like kinase 4
|
|
chr2_+_170336729
|
1.407
|
NM_001110152
|
Pfdn4
|
prefoldin 4
|
|
chr13_-_102462584
|
1.402
|
NM_001024955
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
|