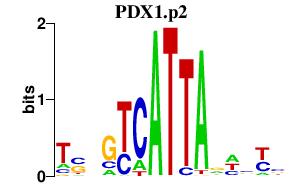
|
chr8_+_56040211
|
18.569
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040553
|
17.373
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040445
|
15.197
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr16_-_34263272
|
3.279
|
|
Kalrn
|
kalirin, RhoGEF kinase
|
|
chr7_-_125676823
|
2.016
|
NM_009940
|
Coq7
|
demethyl-Q 7
|
|
chr7_-_125676764
|
1.822
|
|
Coq7
|
demethyl-Q 7
|
|
chr7_-_125676808
|
1.770
|
|
Coq7
|
demethyl-Q 7
|
|
chr4_+_98697606
|
1.768
|
NM_013913
|
Angptl3
|
angiopoietin-like 3
|
|
chr19_-_40588219
|
1.624
|
NM_178362
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr8_-_42127124
|
1.388
|
|
Mtus1
|
mitochondrial tumor suppressor 1
|
|
chr11_+_31901708
|
1.369
|
|
Nsg2
|
neuron specific gene family member 2
|
|
chr15_-_102455489
|
1.011
|
|
Atf7
|
activating transcription factor 7
|
|
chr2_-_160773804
|
0.753
|
|
Chd6
|
chromodomain helicase DNA binding protein 6
|
|
chr7_+_134123446
|
0.731
|
|
|
|
|
chrX_+_160528118
|
0.703
|
NM_020626
|
Tmem27
|
transmembrane protein 27
|
|
chr9_-_96632093
|
0.672
|
NM_175537
|
Zbtb38
|
zinc finger and BTB domain containing 38
|
|
chr16_+_37580238
|
0.639
|
NM_013547
|
Hgd
|
homogentisate 1, 2-dioxygenase
|
|
chr10_-_126978704
|
0.548
|
|
Lrp1
|
low density lipoprotein receptor-related protein 1
|
|
chr5_+_19413335
|
0.543
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr3_-_107320881
|
0.531
|
NM_172271
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17
|
|
chr1_+_148344816
|
0.520
|
NM_001145807
|
Fam5c
|
family with sequence similarity 5, member C
|
|
chrM_+_11741
|
0.473
|
|
ND5
|
NADH dehydrogenase subunit 5
|
|
chr5_+_138422718
|
0.361
|
NM_013478
|
Azgp1
|
alpha-2-glycoprotein 1, zinc
|
|
chr10_-_67599315
|
0.321
|
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like)
|
|
chr10_+_23614493
|
0.314
|
NM_011704
|
Vnn1
|
vanin 1
|
|
chr15_-_102455844
|
0.290
|
NM_146065
|
Atf7
|
activating transcription factor 7
|
|
chr13_-_30632975
|
0.236
|
|
Uqcrfs1
|
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1
|
|
chr3_+_88373199
|
0.227
|
|
Ubqln4
|
ubiquilin 4
|
|
chr10_-_26095736
|
0.175
|
|
L3mbtl3
|
l(3)mbt-like 3 (Drosophila)
|
|
chr15_-_102455518
|
0.165
|
|
Atf7
|
activating transcription factor 7
|
|
chr9_+_47970096
|
0.157
|
NM_172921
|
Fam55d
|
family with sequence similarity 55, member D
|
|
chr17_-_72102224
|
0.152
|
NM_146082
|
BC027072
|
cDNA sequence BC027072
|
|
chr13_-_54789548
|
0.152
|
NM_001146027
|
Rnf44
|
ring finger protein 44
|
|
chr14_+_48283430
|
0.127
|
NM_008477
|
Ktn1
|
kinectin 1
|
|
chr8_-_108092197
|
0.106
|
NM_007427
|
Agrp
|
agouti related protein
|
|
chr1_+_174886898
|
0.077
|
NM_008763
|
Olfr16
|
olfactory receptor 16
|
|
chr1_+_169298077
|
0.075
|
|
Aldh9a1
|
aldehyde dehydrogenase 9, subfamily A1
|
|
chr9_+_39647020
|
0.065
|
NM_146614
|
Olfr971
|
olfactory receptor 971
|
|
chr7_-_20188580
|
0.047
|
|
Clasrp
|
CLK4-associating serine/arginine rich protein
|
|
chr2_+_91050920
|
0.040
|
|
Acp2
|
acid phosphatase 2, lysosomal
|
|
chr8_+_73993084
|
0.007
|
NM_025600
|
Dda1
|
DET1 and DDB1 associated 1
|