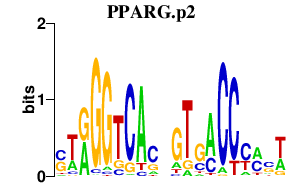
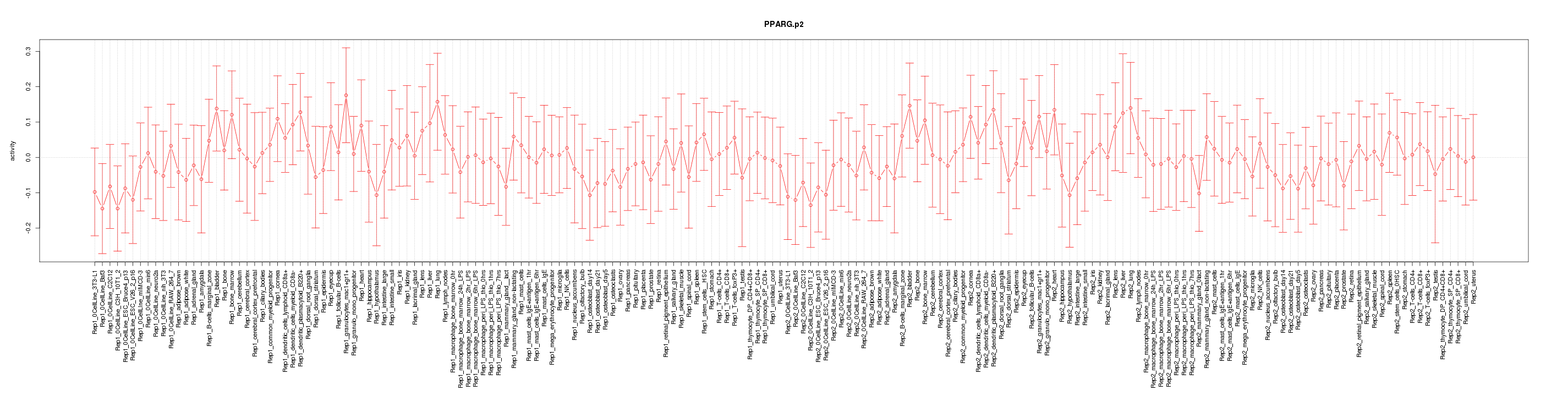
|
chr1_+_133535033
|
7.718
|
|
Ctse
|
cathepsin E
|
|
chr11_-_104330747
|
6.238
|
|
1700081L11Rik
|
RIKEN cDNA 1700081L11 gene
|
|
chr11_+_104445302
|
4.112
|
|
Myl4
|
myosin, light polypeptide 4
|
|
chr7_-_139508857
|
4.083
|
|
Chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr3_-_88258754
|
3.720
|
|
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A
|
|
chr7_-_106289567
|
3.100
|
|
Uvrag
|
UV radiation resistance associated gene
|
|
chr3_+_94282235
|
2.920
|
|
Celf3
|
VCUGBP, Elav-like family member 3
|
|
chr3_+_28596824
|
2.852
|
NM_031197
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2
|
|
chr7_-_106289649
|
2.716
|
|
Uvrag
|
UV radiation resistance associated gene
|
|
chr9_+_110921791
|
2.713
|
NM_008522
|
Ltf
|
lactotransferrin
|
|
chr11_-_50244605
|
2.645
|
NM_172557
|
Rufy1
|
RUN and FYVE domain containing 1
|
|
chr8_+_4238862
|
2.560
|
|
Map2k7
|
mitogen-activated protein kinase kinase 7
|
|
chr17_+_31814873
|
2.406
|
NM_013501
|
Cryaa
|
crystallin, alpha A
|
|
chr11_-_106210699
|
2.373
|
NM_133199
|
Scn4a
|
sodium channel, voltage-gated, type IV, alpha
|
|
chr4_-_41045132
|
2.252
|
NM_016689
|
Aqp3
|
aquaporin 3
|
|
chr6_+_67216948
|
2.131
|
NM_001113564
NM_001113565
NM_001113566
NM_025814
|
Serbp1
|
serpine1 mRNA binding protein 1
|
|
chr14_-_64035873
|
2.114
|
NM_007549
|
Blk
|
B lymphoid kinase
|
|
chr18_-_61946264
|
2.084
|
|
Afap1l1
|
actin filament associated protein 1-like 1
|
|
chr7_-_106289651
|
2.010
|
NM_178635
|
Uvrag
|
UV radiation resistance associated gene
|
|
chr7_-_106289639
|
1.964
|
|
Uvrag
|
UV radiation resistance associated gene
|
|
chr7_-_106289616
|
1.938
|
|
Uvrag
|
UV radiation resistance associated gene
|
|
chr2_+_154016550
|
1.888
|
NM_001012392
|
U46068
|
cDNA sequence U46068
|
|
chr9_+_110322311
|
1.878
|
NM_008694
|
Ngp
|
neutrophilic granule protein
|
|
chr7_-_139508762
|
1.839
|
NM_029935
|
Chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr5_-_146821173
|
1.760
|
NM_019792
|
Cyp3a25
|
cytochrome P450, family 3, subfamily a, polypeptide 25
|
|
chr7_+_29581063
|
1.653
|
|
Rinl
|
Ras and Rab interactor-like
|
|
chr15_-_82624668
|
1.641
|
NM_029562
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26
|
|
chr18_-_61946299
|
1.554
|
NM_178928
|
Afap1l1
|
actin filament associated protein 1-like 1
|
|
chr11_-_69635657
|
1.535
|
NM_028855
|
Spem1
|
sperm maturation 1
|
|
chr17_-_36169460
|
1.468
|
|
|
|
|
chr17_-_36423348
|
1.352
|
NM_177923
|
H2-M10.2
|
histocompatibility 2, M region locus 10.2
|
|
chr6_-_87740264
|
1.336
|
|
Rab43
|
RAB43, member RAS oncogene family
|
|
chr19_-_4190836
|
1.329
|
NM_178650
|
Tbc1d10c
|
TBC1 domain family, member 10c
|
|
chr16_+_4037100
|
1.297
|
NM_010061
|
Dnase1
|
deoxyribonuclease I
|
|
chr12_+_114390258
|
1.289
|
|
Crip1
|
cysteine-rich protein 1 (intestinal)
|
|
chr15_-_82624638
|
1.277
|
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26
|
|
chr1_+_6204729
|
1.256
|
NM_009826
|
Rb1cc1
|
RB1-inducible coiled-coil 1
|
|
chr2_-_126969008
|
1.212
|
NM_001163527
|
Itpripl1
|
inositol 1,4,5-triphosphate receptor interacting protein-like 1
|
|
chr2_+_92215590
|
1.209
|
|
Pex16
|
peroxisomal biogenesis factor 16
|
|
chr1_+_133763777
|
1.133
|
NM_144875
|
Rab7l1
|
RAB7, member RAS oncogene family-like 1
|
|
chr11_-_115992196
|
1.089
|
NM_178802
|
Trim65
|
tripartite motif-containing 65
|
|
chr6_+_113433408
|
1.080
|
|
Creld1
|
cysteine-rich with EGF-like domains 1
|
|
chr6_-_55124983
|
1.070
|
NM_009349
|
Inmt
|
indolethylamine N-methyltransferase
|
|
chr5_-_121227617
|
1.041
|
NM_145226
|
Oas3
|
2'-5' oligoadenylate synthetase 3
|
|
chr19_-_5468482
|
1.036
|
NM_009985
|
Ctsw
|
cathepsin W
|
|
chr12_+_114390222
|
1.035
|
NM_007763
|
Crip1
|
cysteine-rich protein 1 (intestinal)
|
|
chr12_+_86814822
|
1.019
|
NM_010234
|
Fos
|
FBJ osteosarcoma oncogene
|
|
chr8_+_4238838
|
1.010
|
|
Map2k7
|
mitogen-activated protein kinase kinase 7
|
|
chr7_-_135137189
|
0.964
|
|
Pycard
|
PYD and CARD domain containing
|
|
chr5_+_74591290
|
0.960
|
|
Rasl11b
|
RAS-like, family 11, member B
|
|
chr10_-_75227029
|
0.946
|
NM_172549
|
Cabin1
|
calcineurin binding protein 1
|
|
chr6_+_113433507
|
0.943
|
NM_133930
|
Creld1
|
cysteine-rich with EGF-like domains 1
|
|
chr15_-_101543318
|
0.924
|
NM_027011
|
Krt5
|
keratin 5
|
|
chr3_+_87161790
|
0.905
|
NM_009690
|
Cd5l
|
CD5 antigen-like
|
|
chr4_+_43740152
|
0.897
|
|
Hrct1
|
histidine rich carboxyl terminus 1
|
|
chr4_-_63064369
|
0.888
|
NM_001045514
|
Akna
|
AT-hook transcription factor
|
|
chr17_+_34290108
|
0.886
|
NM_010387
|
H2-DMb1
|
histocompatibility 2, class II, locus Mb1
|
|
chr11_+_102297838
|
0.872
|
|
|
|
|
chr5_+_74591336
|
0.864
|
NM_026878
|
Rasl11b
|
RAS-like, family 11, member B
|
|
chr15_-_36485294
|
0.846
|
NM_025712
|
Snx31
|
sorting nexin 31
|
|
chr6_+_70165393
|
0.846
|
|
Igk-V28
Igkv6-23
LOC672450
|
immunoglobulin kappa chain variable 28 (V28)
immunoglobulin kappa variable 6-23
V(kappa) gene product
|
|
chr8_+_4238780
|
0.824
|
|
Map2k7
|
mitogen-activated protein kinase kinase 7
|
|
chr8_+_4238718
|
0.801
|
NM_001042557
NM_001164172
NM_011944
|
Map2k7
|
mitogen-activated protein kinase kinase 7
|
|
chr14_-_56704033
|
0.792
|
NM_008572
|
Mcpt8
|
mast cell protease 8
|
|
chr16_-_4419805
|
0.783
|
|
Adcy9
|
adenylate cyclase 9
|
|
chr7_-_16844888
|
0.725
|
NM_007577
|
C5ar1
|
complement component 5a receptor 1
|
|
chr1_+_173489323
|
0.721
|
NM_018729
|
Cd244
LOC677008
|
CD244 natural killer cell receptor 2B4
natural killer cell receptor 2B4-like
|
|
chr5_+_114185982
|
0.692
|
|
Ficd
|
FIC domain containing
|
|
chr4_+_140462254
|
0.648
|
NM_008812
|
Padi2
|
peptidyl arginine deiminase, type II
|
|
chr11_+_61022240
|
0.634
|
NM_001112725
NM_007436
|
Aldh3a1
|
aldehyde dehydrogenase family 3, subfamily A1
|
|
chr11_+_75007300
|
0.619
|
|
Rtn4rl1
|
reticulon 4 receptor-like 1
|
|
chr4_+_106406496
|
0.611
|
NM_146149
|
Fam151a
|
family with sequence simliarity 151, member A
|
|
chr12_-_70576533
|
0.609
|
|
5830428M24Rik
|
RIKEN cDNA 5830428M24 gene
|
|
chr15_+_82129383
|
0.600
|
NM_029066
|
Wbp2nl
|
WBP2 N-terminal like
|
|
chr6_+_48791481
|
0.593
|
NM_025326
|
Tmem176a
|
transmembrane protein 176A
|
|
chr12_-_55304834
|
0.571
|
NM_028133
|
Egln3
|
EGL nine homolog 3 (C. elegans)
|
|
chr8_+_32239179
|
0.564
|
|
Rnf122
|
ring finger protein 122
|
|
chr6_+_48791616
|
0.553
|
NM_001098271
|
Tmem176a
|
transmembrane protein 176A
|
|
chr16_+_33185137
|
0.504
|
NM_176840
|
Osbpl11
|
oxysterol binding protein-like 11
|
|
chr6_+_112409684
|
0.501
|
|
Cav3
|
caveolin 3
|
|
chr10_-_75226917
|
0.492
|
|
Cabin1
|
calcineurin binding protein 1
|
|
chr9_-_45012097
|
0.491
|
NM_145403
|
Tmprss4
|
transmembrane protease, serine 4
|
|
chr7_-_87884693
|
0.463
|
|
Iqgap1
|
IQ motif containing GTPase activating protein 1
|
|
chr3_+_118265066
|
0.456
|
NM_170778
|
Dpyd
|
dihydropyrimidine dehydrogenase
|
|
chr10_+_119645800
|
0.454
|
NM_026617
|
Tmbim4
|
transmembrane BAX inhibitor motif containing 4
|
|
chr17_-_57367542
|
0.451
|
NM_009778
|
C3
|
complement component 3
|
|
chr11_-_118109789
|
0.448
|
NM_001112699
NM_011180
|
Cyth1
|
cytohesin 1
|
|
chr19_-_61304260
|
0.443
|
NM_009970
|
Csf2ra
|
colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage)
|
|
chr5_+_138082700
|
0.427
|
NM_001081962
|
Sap25
|
sin3 associated polypeptide
|
|
chr4_-_19636000
|
0.414
|
NM_177327
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1
|
|
chr14_-_55585412
|
0.413
|
NM_001164171
NM_010856
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha
|
|
chr2_+_92215395
|
0.402
|
NM_145122
|
Pex16
|
peroxisomal biogenesis factor 16
|
|
chr7_+_148133054
|
0.401
|
|
Athl1
|
ATH1, acid trehalase-like 1 (yeast)
|
|
chr3_-_37211319
|
0.383
|
NM_145825
|
Cetn4
|
centrin 4
|
|
chr5_+_38126757
|
0.382
|
NM_001081106
|
Cytl1
|
cytokine-like 1
|
|
chr4_-_59796621
|
0.382
|
NM_001013577
|
1110054O05Rik
|
RIKEN cDNA 1110054O05 gene
|
|
chr15_+_80502276
|
0.377
|
NM_145986
|
Fam83f
|
family with sequence similarity 83, member F
|
|
chr10_+_127896244
|
0.369
|
NM_001114096
NM_001114097
NM_198160
|
Smarcc2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2
|
|
chr12_-_114099102
|
0.368
|
NM_019925
|
Gpr132
|
G protein-coupled receptor 132
|
|
chr8_+_64406883
|
0.366
|
NM_001081215
|
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60
|
|
chr5_+_3543815
|
0.360
|
NM_001042501
|
Fam133b
|
family with sequence similarity 133, member B
|
|
chr7_-_16844686
|
0.351
|
NM_001173550
|
C5ar1
|
complement component 5a receptor 1
|
|
chr19_-_41876509
|
0.317
|
NM_001163495
NM_027667
|
Arhgap19
|
Rho GTPase activating protein 19
|
|
chr6_-_69193672
|
0.316
|
|
|
|
|
chr5_+_143419271
|
0.313
|
NM_145601
|
Cngb1
|
cyclic nucleotide gated channel beta 1
|
|
chr3_-_36589048
|
0.311
|
NM_019510
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3
|
|
chr14_-_55585407
|
0.304
|
|
Myh6
|
myosin, heavy polypeptide 6, cardiac muscle, alpha
|
|
chr5_+_114185777
|
0.302
|
NM_001010825
|
Ficd
|
FIC domain containing
|
|
chr15_-_102019373
|
0.298
|
|
Csad
|
cysteine sulfinic acid decarboxylase
|
|
chr2_+_172805342
|
0.298
|
NM_001083959
NM_001083960
NM_012046
|
Spo11
|
sporulation protein, meiosis-specific, SPO11 homolog (S. cerevisiae)
|
|
chr1_+_83156336
|
0.295
|
NM_027725
|
Wdr69
|
WD repeat domain 69
|
|
chr5_+_67698177
|
0.288
|
NM_178651
|
Slc30a9
|
solute carrier family 30 (zinc transporter), member 9
|
|
chr6_+_113433656
|
0.286
|
|
Creld1
|
cysteine-rich with EGF-like domains 1
|
|
chr17_-_56615784
|
0.281
|
|
Ptprs
|
protein tyrosine phosphatase, receptor type, S
|
|
chr16_+_87354429
|
0.276
|
NM_001159331
NM_026366
|
N6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative)
|
|
chr11_-_87639611
|
0.270
|
NM_080420
|
Lpo
|
lactoperoxidase
|
|
chr11_+_31865591
|
0.265
|
NM_026262
|
4930524B15Rik
|
RIKEN cDNA 4930524B15 gene
|
|
chr11_+_115773633
|
0.264
|
|
1110017F19Rik
|
RIKEN cDNA 1110017F19 gene
|
|
chr17_+_34093007
|
0.263
|
|
Vps52
|
vacuolar protein sorting 52 (yeast)
|
|
chr3_+_137976249
|
0.260
|
NM_026945
|
Adh6a
|
alcohol dehydrogenase 6A (class V)
|
|
chrX_+_154137049
|
0.259
|
NM_025357
|
Smpx
|
small muscle protein, X-linked
|
|
chr10_-_79811127
|
0.258
|
NM_025629
NM_001113548
|
Adamtsl5
|
ADAMTS-like 5
|
|
chr14_-_55613384
|
0.256
|
NM_080728
|
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta
|
|
chr18_-_34784463
|
0.250
|
NM_030147
|
Brd8
|
bromodomain containing 8
|
|
chr5_-_115108377
|
0.249
|
NM_022017
|
Trpv4
|
transient receptor potential cation channel, subfamily V, member 4
|
|
chr7_+_86418127
|
0.233
|
NM_018811
|
Abhd2
|
abhydrolase domain containing 2
|
|
chr7_-_108437135
|
0.226
|
|
|
|
|
chr10_+_57834201
|
0.224
|
NM_201242
|
Lims1
|
LIM and senescent cell antigen-like domains 1
|
|
chr8_-_97830469
|
0.223
|
NM_001195413
NM_145601
|
Cngb1
|
cyclic nucleotide gated channel beta 1
|
|
chr5_+_90947970
|
0.216
|
NM_145146
|
Afm
|
afamin
|
|
chr11_+_33863012
|
0.213
|
NM_031169
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1
|
|
chr3_-_33041993
|
0.209
|
NM_001163516
|
Pex5l
|
peroxisomal biogenesis factor 5-like
|
|
chr10_+_59764990
|
0.204
|
|
Psap
|
prosaposin
|
|
chr12_+_88607621
|
0.204
|
NM_146036
|
Ahsa1
|
AHA1, activator of heat shock protein ATPase homolog 1 (yeast)
|
|
chr7_+_13551155
|
0.192
|
|
Zfp324
|
zinc finger protein 324
|
|
chr5_+_124380677
|
0.190
|
NM_001134767
|
Ccdc62
|
coiled-coil domain containing 62
|
|
chr17_-_83913636
|
0.180
|
NM_001159529
NM_009187
|
Cox7a2l
|
cytochrome c oxidase subunit VIIa polypeptide 2-like
|
|
chrX_-_151467834
|
0.171
|
NM_020016
|
Magea2
|
melanoma antigen, family A, 2
|
|
chr9_+_45211220
|
0.155
|
NM_052823
|
Fxyd2
|
FXYD domain-containing ion transport regulator 2
|
|
chr13_-_34437042
|
0.153
|
NM_001033167
|
Slc22a23
|
solute carrier family 22, member 23
|
|
chr6_+_120786323
|
0.151
|
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator)
|
|
chr7_-_110809831
|
0.138
|
NM_147122
|
Olfr623
|
olfactory receptor 623
|
|
chr3_-_33042050
|
0.131
|
|
Pex5l
|
peroxisomal biogenesis factor 5-like
|
|
chr2_+_74513069
|
0.118
|
NM_008274
|
Hoxd12
|
homeobox D12
|
|
chr6_+_112409498
|
0.109
|
NM_007617
|
Cav3
|
caveolin 3
|
|
chr5_+_138187182
|
0.104
|
NM_023910
|
Tsc22d4
|
TSC22 domain family, member 4
|
|
chr19_-_3897078
|
0.104
|
|
Tcirg1
|
T-cell, immune regulator 1, ATPase, H+ transporting, lysosomal V0 protein A3
|
|
chr7_-_109907908
|
0.095
|
NM_001168503
NM_130866
|
Olfr78
|
olfactory receptor 78
|
|
chr8_+_122638031
|
0.093
|
NM_001163761
NM_001163762
NM_172286
|
6430548M08Rik
|
RIKEN cDNA 6430548M08 gene
|
|
chr8_+_63702967
|
0.090
|
NM_021506
|
Sh3rf1
|
SH3 domain containing ring finger 1
|
|
chr8_-_107813357
|
0.090
|
NM_001166394
NM_028888
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene
|
|
chr14_+_61791811
|
0.085
|
|
Sacs
|
sacsin
|
|
chr11_-_69011310
|
0.082
|
NM_009661
|
Alox8
|
arachidonate 8-lipoxygenase
|
|
chr7_-_134592467
|
0.081
|
NM_175163
|
Zfp689
|
zinc finger protein 689
|
|
chr7_+_13551210
|
0.079
|
NM_178732
|
Zfp324
|
zinc finger protein 324
|
|
chr2_-_155127037
|
0.073
|
|
Pigu
|
phosphatidylinositol glycan anchor biosynthesis, class U
|
|
chr11_-_97637190
|
0.068
|
NM_028156
|
1700001P01Rik
|
RIKEN cDNA 1700001P01 gene
|
|
chr5_+_149818706
|
0.066
|
|
LOC100503307
|
hypothetical LOC100503307
|
|
chr8_-_75005707
|
0.058
|
|
Slc35e1
|
solute carrier family 35, member E1
|
|
chr17_+_35204268
|
0.056
|
NM_023463
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C
|
|
chr1_+_75143987
|
0.033
|
|
Fam134a
|
family with sequence similarity 134, member A
|
|
chr4_+_108000728
|
0.033
|
NM_027250
|
2010305A19Rik
|
RIKEN cDNA 2010305A19 gene
|
|
chr10_-_7400836
|
0.032
|
|
BC020402
|
cDNA sequence BC020402
|
|
chr7_+_134592155
|
0.017
|
|
B130055M24Rik
|
RIKEN cDNA B130055M24 gene
|
|
chr9_-_55359898
|
0.012
|
|
Etfa
|
electron transferring flavoprotein, alpha polypeptide
|
|
chr6_-_52114644
|
0.010
|
NM_010451
|
Hoxa2
|
homeobox A2
|
|
chr18_-_38498564
|
0.010
|
NM_011937
|
Gnpda1
|
glucosamine-6-phosphate deaminase 1
|