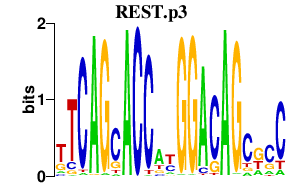
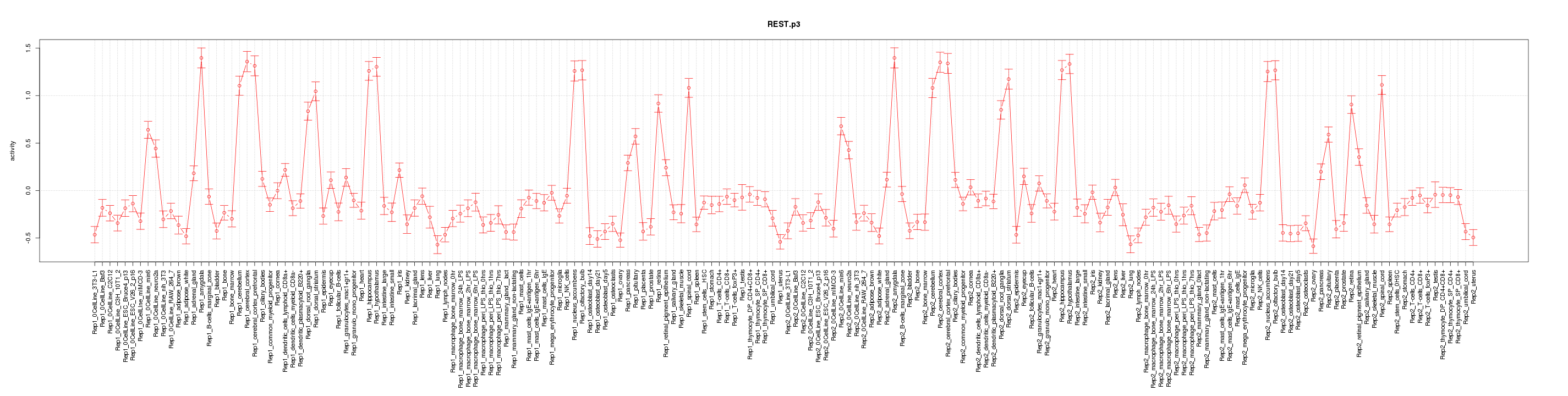
|
chr2_+_132607432
|
274.238
|
|
Chgb
|
chromogranin B
|
|
chr2_+_132607013
|
274.060
|
NM_007694
|
Chgb
|
chromogranin B
|
|
chr2_+_132607398
|
269.078
|
|
Chgb
|
chromogranin B
|
|
chr2_+_132607408
|
267.647
|
|
Chgb
|
chromogranin B
|
|
chr2_-_181049204
|
260.292
|
NM_009133
|
Stmn3
|
stathmin-like 3
|
|
chr2_-_181048957
|
254.386
|
|
Stmn3
|
stathmin-like 3
|
|
chr16_-_23890909
|
243.530
|
NM_009215
|
Sst
|
somatostatin
|
|
chr6_-_121423633
|
223.441
|
|
Iqsec3
|
IQ motif and Sec7 domain 3
|
|
chrX_+_7215705
|
214.562
|
NM_009305
|
Syp
|
synaptophysin
|
|
chr18_-_31606908
|
203.883
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chr3_-_80606500
|
201.906
|
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr2_+_164793698
|
198.509
|
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr1_-_84692750
|
191.074
|
NM_152915
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chr6_-_121423682
|
190.570
|
NM_001033354
|
Iqsec3
|
IQ motif and Sec7 domain 3
|
|
chr12_+_103793130
|
188.428
|
NM_007693
|
Chga
|
chromogranin A
|
|
chr4_-_128798481
|
187.735
|
NM_010471
|
Hpca
|
hippocalcin
|
|
chr1_-_84692352
|
185.430
|
|
Dner
|
delta/notch-like EGF-related receptor
|
|
chrX_-_20498021
|
174.139
|
NM_001110780
NM_013680
|
Syn1
|
synapsin I
|
|
chr11_-_35612385
|
165.999
|
NM_001004147
|
Fbll1
|
fibrillarin-like 1
|
|
chr5_+_137506122
|
157.171
|
NM_001039385
|
Vgf
|
VGF nerve growth factor inducible
|
|
chr3_-_80606704
|
154.941
|
NM_001039195
NM_001083806
NM_013540
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr5_-_121459496
|
145.759
|
NM_011286
|
Rph3a
|
rabphilin 3A
|
|
chr1_-_177422630
|
145.460
|
NM_001199003
NM_011880
|
Rgs7
|
regulator of G protein signaling 7
|
|
chr4_+_101223198
|
145.049
|
NM_001164583
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr11_-_35612058
|
144.365
|
|
Fbll1
|
fibrillarin-like 1
|
|
chr12_-_113749361
|
143.847
|
|
Tmem179
|
transmembrane protein 179
|
|
chr3_-_80606641
|
139.985
|
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr7_+_28438881
|
139.830
|
NM_028417
|
Ttc9b
|
tetratricopeptide repeat domain 9B
|
|
chr12_-_113749313
|
137.121
|
|
Tmem179
|
transmembrane protein 179
|
|
chr11_+_102254949
|
134.568
|
|
Rundc3a
|
RUN domain containing 3A
|
|
chr11_+_102254816
|
133.191
|
|
Rundc3a
|
RUN domain containing 3A
|
|
chr12_-_113749363
|
128.899
|
NM_178915
|
Tmem179
|
transmembrane protein 179
|
|
chr2_+_164793796
|
125.235
|
|
Slc12a5
|
solute carrier family 12, member 5
|
|
chr4_-_4065205
|
123.610
|
|
Penk
|
preproenkephalin
|
|
chr2_-_92241282
|
122.977
|
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr2_+_22478062
|
116.323
|
|
Gad2
|
glutamic acid decarboxylase 2
|
|
chr2_-_92241357
|
114.221
|
NM_011162
|
Mapk8ip1
|
mitogen-activated protein kinase 8 interacting protein 1
|
|
chr2_+_22478131
|
113.265
|
|
Gad2
|
glutamic acid decarboxylase 2
|
|
chr11_+_102254699
|
113.122
|
NM_016759
|
Rundc3a
|
RUN domain containing 3A
|
|
chr2_-_25174498
|
111.242
|
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1)
|
|
chr3_+_67868723
|
107.287
|
NM_001113420
NM_001113421
|
Schip1
|
schwannomin interacting protein 1
|
|
chr4_+_15808409
|
106.684
|
NM_009788
|
Calb1
|
calbindin 1
|
|
chr12_-_71448300
|
103.462
|
|
Trim9
|
tripartite motif-containing 9
|
|
chr7_+_58877199
|
100.479
|
NM_080853
|
Slc17a6
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6
|
|
chr2_-_25174662
|
97.670
|
NM_001177656
NM_001177657
NM_008169
|
Grin1
|
glutamate receptor, ionotropic, NMDA1 (zeta 1)
|
|
chr13_-_100670595
|
95.890
|
NM_001081493
NM_013732
|
Cartpt
|
CART prepropeptide
|
|
chr9_+_104472090
|
95.534
|
NM_028719
|
Cpne4
|
copine IV
|
|
chr7_-_88638789
|
94.668
|
NM_021492
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr2_+_22477837
|
94.430
|
NM_008078
|
Gad2
|
glutamic acid decarboxylase 2
|
|
chr5_-_121459315
|
89.814
|
|
Rph3a
|
rabphilin 3A
|
|
chr7_-_28181562
|
89.301
|
|
Spnb4
|
spectrin beta 4
|
|
chr9_+_40076800
|
86.266
|
NM_001083917
NM_153522
NM_178227
|
Scn3b
|
sodium channel, voltage-gated, type III, beta
|
|
chr9_+_108728650
|
84.417
|
NM_080437
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila)
|
|
chr15_-_76552594
|
83.530
|
NM_198119
|
Lrrc24
|
leucine rich repeat containing 24
|
|
chr11_+_80290571
|
82.418
|
|
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr6_+_8898613
|
81.103
|
|
Nxph1
|
neurexophilin 1
|
|
chr16_-_37385024
|
72.609
|
NM_001114611
NM_001114612
NM_001114613
NM_172440
|
Stxbp5l
|
syntaxin binding protein 5-like
|
|
chr9_-_57315800
|
72.391
|
NM_020270
|
Scamp5
|
secretory carrier membrane protein 5
|
|
chr10_-_4707228
|
71.574
|
NM_011702
|
Vip
|
vasoactive intestinal polypeptide
|
|
chr10_+_90292388
|
68.670
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr1_-_177422520
|
68.216
|
|
Rgs7
|
regulator of G protein signaling 7
|
|
chr12_-_71448600
|
65.838
|
NM_001110202
NM_001110203
NM_053167
|
Trim9
|
tripartite motif-containing 9
|
|
chr6_+_113232319
|
65.576
|
|
Cpne9
|
copine family member IX
|
|
chr6_-_134837738
|
65.454
|
NM_001167697
NM_001167699
NM_008157
|
Gpr19
|
G protein-coupled receptor 19
|
|
chr11_+_80290509
|
64.726
|
NM_009871
|
Cdk5r1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35)
|
|
chr13_+_54472705
|
58.851
|
NM_009946
|
Cplx2
|
complexin 2
|
|
chr2_-_79294977
|
56.825
|
|
Neurod1
|
neurogenic differentiation 1
|
|
chr7_+_70589623
|
52.566
|
NM_130880
|
Otud7a
|
OTU domain containing 7A
|
|
chr1_+_34636537
|
51.751
|
NM_213727
|
Fam123c
|
family with sequence similarity 123, member C
|
|
chr11_+_77744280
|
50.466
|
NM_021286
|
Sez6
|
seizure related gene 6
|
|
chr4_+_12833983
|
49.328
|
NM_001171801
NM_173746
|
Gm11818
|
predicted gene 11818
|
|
chr2_+_25719373
|
49.316
|
NM_001145403
|
Kcnt1
|
potassium channel, subfamily T, member 1
|
|
chr6_+_113232300
|
47.852
|
NM_170673
|
Cpne9
|
copine family member IX
|
|
chr19_+_42330170
|
41.416
|
NM_027694
|
Golga7b
|
golgi autoantigen, golgin subfamily a, 7B
|
|
chr7_+_108570359
|
40.860
|
|
Pde2a
|
phosphodiesterase 2A, cGMP-stimulated
|
|
chr13_-_105844882
|
40.647
|
NM_029879
|
Rgs7bp
|
regulator of G-protein signalling 7 binding protein
|
|
chr17_-_91492065
|
40.513
|
NM_020252
NM_177284
|
Nrxn1
|
neurexin I
|
|
chr12_-_86039703
|
38.872
|
NM_001033334
|
Tmem90a
|
transmembrane protein 90a
|
|
chr7_+_26102182
|
38.255
|
NM_001160400
|
Megf8
|
multiple EGF-like-domains 8
|
|
chr12_+_117724182
|
35.154
|
NM_011215
|
Ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr17_-_25377052
|
34.871
|
NM_172529
|
Gnptg
|
N-acetylglucosamine-1-phosphotransferase, gamma subunit
|
|
chr17_+_70318387
|
33.357
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr14_+_66587319
|
31.697
|
NM_013492
|
Clu
|
clusterin
|
|
chr7_-_88638540
|
30.788
|
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit
|
|
chr9_+_59426108
|
30.750
|
NM_175235
|
Celf6
|
CUGBP, Elav-like family member 6
|
|
chr3_+_94282235
|
29.854
|
|
Celf3
|
VCUGBP, Elav-like family member 3
|
|
chr17_+_70318530
|
27.109
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr17_-_23877115
|
25.957
|
NM_023824
|
Paqr4
|
progestin and adipoQ receptor family member IV
|
|
chr3_+_114606995
|
25.192
|
NM_153458
|
Olfm3
|
olfactomedin 3
|
|
chr13_-_54850954
|
24.462
|
NM_012014
|
Gprin1
|
G protein-regulated inducer of neurite outgrowth 1
|
|
chr10_+_90292453
|
24.416
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr3_-_89568552
|
23.296
|
NM_009602
|
Chrnb2
|
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal)
|
|
chr2_-_179839111
|
23.282
|
|
Hrh3
|
histamine receptor H3
|
|
chr2_-_179838878
|
22.485
|
NM_133849
|
Hrh3
|
histamine receptor H3
|
|
chr4_+_128814102
|
21.348
|
NM_027402
|
Fndc5
|
fibronectin type III domain containing 5
|
|
chr17_+_28712663
|
19.473
|
NM_026571
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5
|
|
chr13_+_45602853
|
18.675
|
|
Gmpr
|
guanosine monophosphate reductase
|
|
chr3_-_89568298
|
17.518
|
|
Chrnb2
|
cholinergic receptor, nicotinic, beta polypeptide 2 (neuronal)
|
|
chr2_+_139996419
|
17.193
|
|
2310003L22Rik
|
RIKEN cDNA 2310003L22 gene
|
|
chr2_+_139996381
|
15.742
|
NM_027093
|
2310003L22Rik
|
RIKEN cDNA 2310003L22 gene
|
|
chr4_-_152856922
|
13.912
|
NM_001099299
|
Ajap1
|
adherens junction associated protein 1
|
|
chr13_+_45602825
|
13.500
|
NM_025508
|
Gmpr
|
guanosine monophosphate reductase
|
|
chr11_+_58144207
|
11.746
|
NM_001161338
NM_024480
|
Sh3bp5l
|
SH3 binding domain protein 5 like
|
|
chr4_-_135050358
|
10.782
|
NM_028995
|
Nipal3
|
NIPA-like domain containing 3
|
|
chr17_+_70318707
|
10.666
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr9_+_122026424
|
10.131
|
|
Snrk
|
SNF related kinase
|
|
chr7_-_20189709
|
10.109
|
NM_016680
|
Clasrp
|
CLK4-associating serine/arginine rich protein
|
|
chr13_+_45602909
|
9.972
|
|
Gmpr
|
guanosine monophosphate reductase
|
|
chr11_+_74433092
|
7.599
|
NM_001013784
|
E130309D14Rik
|
RIKEN cDNA E130309D14 gene
|
|
chr8_+_18846262
|
7.579
|
NM_026792
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr8_-_69211936
|
7.288
|
NM_016708
|
Npy5r
|
neuropeptide Y receptor Y5
|
|
chr16_-_17576750
|
7.100
|
NM_144852
|
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4
|
|
chr17_-_71339826
|
6.820
|
NM_023402
|
Myl12b
|
myosin, light chain 12B, regulatory
|
|
chr19_+_5038825
|
6.131
|
NM_175383
|
B3gnt1
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1
|
|
chr2_-_26997965
|
5.271
|
NM_175427
|
Fam163b
|
family with sequence similarity 163, member B
|
|
chr15_+_101303908
|
4.584
|
NM_010667
|
Krt86
|
keratin 86
|
|
chrX_-_100103981
|
4.405
|
NM_027591
|
Dmrtc1a
|
DMRT-like family C1a
|
|
chr12_+_117724251
|
4.318
|
|
Ptprn2
|
protein tyrosine phosphatase, receptor type, N polypeptide 2
|
|
chr11_-_115465172
|
4.188
|
NM_173048
|
Gga3
|
golgi associated, gamma adaptin ear containing, ARF binding protein 3
|
|
chr17_-_46841925
|
3.988
|
NM_009358
|
Ppp2r5d
|
protein phosphatase 2, regulatory subunit B (B56), delta isoform
|
|
chr16_-_45742995
|
3.374
|
NM_172511
|
Abhd10
|
abhydrolase domain containing 10
|
|
chr2_+_174123359
|
3.065
|
NM_001077507
NM_010309
NM_201617
NM_201618
|
Gnas
|
GNAS (guanine nucleotide binding protein, alpha stimulating) complex locus
|
|
chr9_-_21889810
|
3.057
|
NM_012029
|
Ecsit
|
ECSIT homolog (Drosophila)
|
|
chr19_+_47253484
|
2.984
|
|
Neurl1a
|
neuralized homolog 1A (Drosophila)
|
|
chr11_-_74710462
|
2.463
|
|
Sgsm2
|
small G protein signaling modulator 2
|
|
chr19_+_47253309
|
1.822
|
NM_021360
|
Neurl1a
|
neuralized homolog 1A (Drosophila)
|
|
chr12_+_114378688
|
1.612
|
|
Crip2
|
cysteine rich protein 2
|
|
chr8_+_18846471
|
1.340
|
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|
|
chr8_-_13253995
|
1.328
|
NM_172750
|
Adprhl1
|
ADP-ribosylhydrolase like 1
|
|
chr9_-_21889841
|
1.187
|
|
Ecsit
|
ECSIT homolog (Drosophila)
|
|
chr1_-_164478628
|
0.827
|
|
Mettl13
|
methyltransferase like 13
|
|
chr19_+_6334978
|
0.743
|
NM_001168488
NM_001168489
NM_001168490
NM_008583
|
Men1
|
multiple endocrine neoplasia 1
|
|
chr17_+_69788665
|
0.651
|
NM_001145192
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene
|
|
chr5_+_117570113
|
0.648
|
|
Taok3
|
TAO kinase 3
|
|
chr8_+_18846524
|
0.300
|
|
Agpat5
|
1-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon)
|