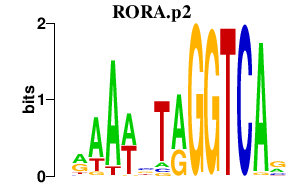
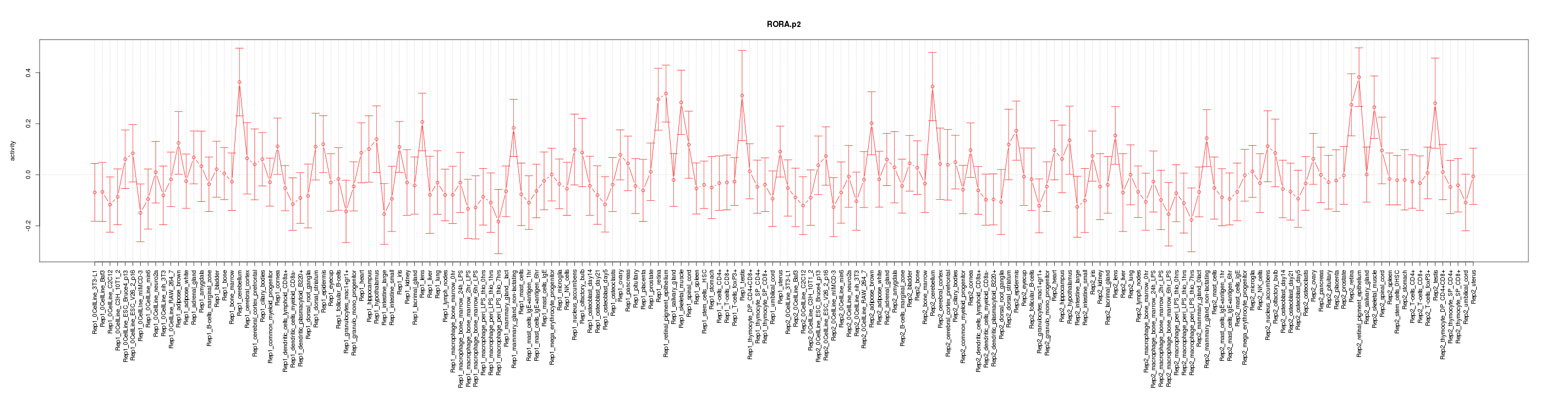
|
chr19_+_6384413
|
26.418
|
NM_011224
|
Pygm
|
muscle glycogen phosphorylase
|
|
chr9_-_53822931
|
26.393
|
|
Elmod1
|
ELMO domain containing 1
|
|
chr9_-_53823107
|
24.916
|
NM_177769
|
Elmod1
|
ELMO domain containing 1
|
|
chr7_+_52960585
|
24.414
|
NM_016974
|
Dbp
|
D site albumin promoter binding protein
|
|
chr7_+_52960702
|
20.601
|
|
Dbp
|
D site albumin promoter binding protein
|
|
chr11_-_69418965
|
16.018
|
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr18_+_23468585
|
12.434
|
|
Dtna
|
dystrobrevin alpha
|
|
chr9_-_86589782
|
11.179
|
NM_008615
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr12_-_82434150
|
10.884
|
NM_001167920
NM_080440
|
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3
|
|
chr14_-_56143799
|
10.706
|
NM_001136074
|
Nrl
|
neural retina leucine zipper gene
|
|
chr15_-_7348093
|
10.632
|
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr15_-_7348393
|
9.954
|
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr7_+_53693190
|
9.210
|
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1
|
|
chr8_-_3625251
|
7.986
|
NM_008790
|
Pcp2
|
Purkinje cell protein 2 (L7)
|
|
chrX_-_99447501
|
7.907
|
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1
|
|
chr2_+_90725895
|
7.492
|
NM_026161
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4
|
|
chr10_+_56097078
|
7.456
|
NM_010288
|
Gja1
|
gap junction protein, alpha 1
|
|
chr6_+_113408477
|
7.296
|
NM_145826
|
Il17re
|
interleukin 17 receptor E
|
|
chr15_-_7348290
|
6.858
|
NM_178748
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains
|
|
chr10_+_5151833
|
6.809
|
NM_001079686
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr8_-_3624988
|
6.606
|
NM_001129804
|
Pcp2
|
Purkinje cell protein 2 (L7)
|
|
chr1_+_74589461
|
6.062
|
NM_001081456
|
Plcd4
|
phospholipase C, delta 4
|
|
chr7_+_120350963
|
5.952
|
NM_007489
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr6_+_104443037
|
5.917
|
NM_017383
|
Cntn6
|
contactin 6
|
|
chr7_+_120351012
|
5.643
|
|
Arntl
|
aryl hydrocarbon receptor nuclear translocator-like
|
|
chr10_+_79630579
|
5.386
|
NM_007705
|
Cirbp
|
cold inducible RNA binding protein
|
|
chr15_+_99532787
|
5.319
|
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr3_+_146067763
|
5.301
|
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr6_+_90424727
|
4.820
|
|
Klf15
|
Kruppel-like factor 15
|
|
chr15_+_99532698
|
4.702
|
NM_031842
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr12_+_84861415
|
4.699
|
NM_030246
|
Dcaf4
|
DDB1 and CUL4 associated factor 4
|
|
chr6_+_113408650
|
4.680
|
NM_001034029
NM_001034031
|
Il17re
|
interleukin 17 receptor E
|
|
chr6_-_88824037
|
4.561
|
NM_176973
|
Podxl2
|
podocalyxin-like 2
|
|
chr17_+_25023618
|
4.260
|
NM_001163750
NM_001163751
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr5_+_144861227
|
4.219
|
NM_001081109
|
Lmtk2
|
lemur tyrosine kinase 2
|
|
chr9_+_43915391
|
3.788
|
NM_001190319
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5
|
|
chr12_+_118754951
|
3.451
|
NM_175930
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr19_+_23761879
|
3.418
|
NM_028208
|
Ptar1
|
protein prenyltransferase alpha subunit repeat containing 1
|
|
chr11_+_3414884
|
3.349
|
|
Selm
|
selenoprotein M
|
|
chr7_-_35437859
|
3.291
|
NM_146188
|
Kctd15
|
potassium channel tetramerisation domain containing 15
|
|
chr3_-_107734481
|
3.290
|
NM_026672
|
Gstm7
|
glutathione S-transferase, mu 7
|
|
chr15_-_74583382
|
3.269
|
NM_011838
|
Lynx1
|
Ly6/neurotoxin 1
|
|
chr3_+_89195758
|
3.164
|
|
|
|
|
chr7_+_133416177
|
2.989
|
|
Sbk1
|
SH3-binding kinase 1
|
|
chr8_-_3625510
|
2.915
|
NM_001129803
|
Pcp2
|
Purkinje cell protein 2 (L7)
|
|
chr1_-_182123596
|
2.874
|
NM_001163290
|
Adck3
|
aarF domain containing kinase 3
|
|
chr14_+_37881214
|
2.765
|
NM_173418
|
Lrit2
|
leucine-rich repeat, immunoglobulin-like and transmembrane domains 2
|
|
chr6_+_4471231
|
2.696
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr6_-_29330414
|
2.658
|
NM_007538
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan)
|
|
chr7_+_96781322
|
2.614
|
NM_181407
|
Me3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial
|
|
chr14_+_21493519
|
2.544
|
NM_001168273
NM_172596
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr3_+_67868723
|
2.440
|
NM_001113420
NM_001113421
|
Schip1
|
schwannomin interacting protein 1
|
|
chr16_+_41533317
|
2.422
|
NM_175548
|
Lsamp
|
limbic system-associated membrane protein
|
|
chr11_+_121296252
|
2.339
|
NM_001038699
NM_022014
|
Fn3k
|
fructosamine 3 kinase
|
|
chr17_+_24869629
|
2.288
|
NM_025425
|
Rpl3l
|
ribosomal protein L3-like
|
|
chr17_-_79135885
|
2.243
|
NM_011500
|
Strn
|
striatin, calmodulin binding protein
|
|
chr11_-_20729367
|
2.029
|
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr9_+_20673178
|
1.971
|
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene
|
|
chr12_-_16596573
|
1.939
|
NM_001130412
NM_015763
NM_172950
|
Lpin1
|
lipin 1
|
|
chr6_+_34426352
|
1.931
|
NM_007563
|
Bpgm
|
2,3-bisphosphoglycerate mutase
|
|
chr11_-_99146520
|
1.914
|
NM_029393
|
Krt24
|
keratin 24
|
|
chr3_+_137530638
|
1.860
|
NM_001033155
|
Dnajb14
|
DnaJ (Hsp40) homolog, subfamily B, member 14
|
|
chr12_+_53955707
|
1.762
|
|
Akap6
|
A kinase (PRKA) anchor protein 6
|
|
chr11_-_116060026
|
1.499
|
|
Acox1
|
acyl-Coenzyme A oxidase 1, palmitoyl
|
|
chr17_-_24135049
|
1.478
|
NM_133731
|
Prss22
|
protease, serine, 22
|
|
chr9_-_54407955
|
1.451
|
|
Cib2
|
calcium and integrin binding family member 2
|
|
chr8_+_113618490
|
1.444
|
|
Pdpr
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chrX_+_150158113
|
1.440
|
|
2210013O21Rik
|
RIKEN cDNA 2210013O21 gene
|
|
chr1_+_53370483
|
1.417
|
NM_028091
|
Osgepl1
|
O-sialoglycoprotein endopeptidase-like 1
|
|
chr2_+_48805027
|
1.399
|
NM_029924
|
Mbd5
|
methyl-CpG binding domain protein 5
|
|
chr12_-_16596567
|
1.394
|
|
Lpin1
|
lipin 1
|
|
chr9_+_43915327
|
1.360
|
NM_001040631
NM_001040632
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5
|
|
chrX_-_26129763
|
1.309
|
NM_001162364
|
Gm14525
|
predicted gene 14525
|
|
chrX_-_24728580
|
1.309
|
NM_001040669
|
Gm5169
|
predicted gene 5169
|
|
chr18_-_68459753
|
1.296
|
|
4933403F05Rik
|
RIKEN cDNA 4933403F05 gene
|
|
chr14_+_56194510
|
1.292
|
NM_026808
|
Fitm1
|
fat storage-inducing transmembrane protein 1
|
|
chr6_+_124948627
|
1.280
|
NM_001145926
|
C530028O21Rik
|
RIKEN cDNA C530028O21 gene
|
|
chr6_+_48504791
|
1.277
|
NM_001110160
NM_177736
|
Lrrc61
|
leucine rich repeat containing 61
|
|
chr11_+_52202137
|
1.244
|
|
Vdac1
|
voltage-dependent anion channel 1
|
|
chr11_-_115489392
|
1.202
|
NM_026071
|
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19
|
|
chr17_+_33775000
|
1.174
|
NM_001002842
|
Pram1
|
PML-RAR alpha-regulated adaptor molecule 1
|
|
chr17_+_23737822
|
1.156
|
NM_001033425
|
Zscan10
|
zinc finger and SCAN domain containing 10
|
|
chr3_-_157587998
|
1.084
|
NM_145953
|
Cth
|
cystathionase (cystathionine gamma-lyase)
|
|
chr9_+_75289299
|
1.073
|
NM_001039522
|
Leo1
|
Leo1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae)
|
|
chr4_-_138375124
|
1.068
|
NM_011110
|
Pla2g5
|
phospholipase A2, group V
|
|
chr1_+_20720985
|
1.038
|
NM_010552
|
Il17a
|
interleukin 17A
|
|
chr12_+_80309899
|
0.993
|
NM_030017
|
Rdh12
|
retinol dehydrogenase 12
|
|
chrX_-_26362794
|
0.975
|
NM_001104946
NM_001114754
|
Gm2012
Gm6121
|
predicted gene 2012
predicted gene 6121
|
|
chr9_+_77602582
|
0.957
|
NM_010295
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr9_+_20673069
|
0.947
|
NM_175687
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene
|
|
chr7_-_7162599
|
0.858
|
NM_145577
|
Zfp772
|
zinc finger protein 772
|
|
chr11_-_97011025
|
0.855
|
NM_198100
|
Tbkbp1
|
TBK1 binding protein 1
|
|
chr3_+_146067605
|
0.855
|
NM_138744
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein
|
|
chr10_-_120335907
|
0.834
|
NM_177092
|
Msrb3
|
methionine sulfoxide reductase B3
|
|
chr2_+_29821079
|
0.816
|
NM_001076554
NM_001177667
NM_001177668
|
Spna2
|
spectrin alpha 2
|
|
chr18_+_85104396
|
0.807
|
NM_015798
|
Fbxo15
|
F-box protein 15
|
|
chr17_+_88071837
|
0.728
|
NM_008628
|
Msh2
|
mutS homolog 2 (E. coli)
|
|
chr2_+_119851954
|
0.722
|
|
Mapkbp1
|
mitogen-activated protein kinase binding protein 1
|
|
chr14_-_51766415
|
0.714
|
NM_011271
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic)
|
|
chrX_-_26904190
|
0.698
|
NM_001109969
NM_001099919
NM_001102678
NM_001109970
NM_001110250
NM_001100610
NM_001099347
|
Gm10058
Gm10147
Gm10096
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_-_24498453
|
0.698
|
NM_001109969
NM_001099919
NM_001102678
NM_001109970
NM_001110250
NM_001100610
NM_001099347
|
Gm10058
Gm10147
Gm10096
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_+_29130521
|
0.674
|
NM_001109969
NM_001099919
NM_001102678
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
|
Gm10058
Gm10147
Gm10096
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chr10_+_77079550
|
0.668
|
|
|
|
|
chrX_-_27251094
|
0.658
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_-_28258880
|
0.658
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_-_26822007
|
0.646
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_-_27912080
|
0.646
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chr6_+_90554873
|
0.617
|
NM_027868
|
Slc41a3
|
solute carrier family 41, member 3
|
|
chrX_-_29947647
|
0.616
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_-_27597810
|
0.616
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_+_29483950
|
0.606
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_+_32944181
|
0.605
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chrX_+_32607378
|
0.605
|
NM_001109969
NM_001099919
NM_001102678
NM_001100609
NM_001109970
NM_001110250
NM_001099325
NM_001100610
NM_001099347
NM_009529
|
Gm10058
Gm10147
Gm10096
Gm10487
Gm10486
Gm14819
Gm10488
Gm14632
Gm10230
Gm4836
|
predicted gene 10058
predicted gene 10147
predicted gene 10096
predicted gene 10487
predicted gene 10486
predicted gene 14819
predicted gene 10488
predicted gene 14632
predicted gene 10230
predicted gene 4836
|
|
chr7_+_127883556
|
0.605
|
|
Vwa3a
|
von Willebrand factor A domain containing 3A
|
|
chr17_-_31774075
|
0.596
|
NM_178224
NM_144855
|
Cbs
|
cystathionine beta-synthase
|
|
chr15_-_97561769
|
0.530
|
NM_001168693
|
Endou
|
endonuclease, polyU-specific
|
|
chr17_+_72022598
|
0.518
|
NM_177087
|
Fam179a
|
family with sequence similarity 179, member A
|
|
chr9_+_77602377
|
0.517
|
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr4_-_82666973
|
0.513
|
NM_177863
|
Frem1
|
Fras1 related extracellular matrix protein 1
|
|
chr17_+_13200673
|
0.495
|
NM_013671
|
Sod2
|
superoxide dismutase 2, mitochondrial
|
|
chr3_-_72860864
|
0.476
|
NM_198864
|
Slitrk3
|
SLIT and NTRK-like family, member 3
|
|
chr11_+_109405024
|
0.473
|
NM_001166177
|
Arsg
|
arylsulfatase G
|
|
chrX_-_24352289
|
0.466
|
NM_001081657
|
Gm5935
|
predicted gene 5935
|
|
chr14_-_55482884
|
0.407
|
NM_001177705
|
Homez
|
homeodomain leucine zipper-encoding gene
|
|
chr6_+_34362333
|
0.390
|
NM_009731
|
Akr1b7
|
aldo-keto reductase family 1, member B7
|
|
chr11_-_78059124
|
0.389
|
NM_009297
|
Supt6h
|
suppressor of Ty 6 homolog (S. cerevisiae)
|
|
chr15_-_77473707
|
0.375
|
NM_001143686
|
Apol11b
|
apolipoprotein L 11b
|
|
chr13_-_21842373
|
0.359
|
NM_178211
|
Hist1h4k
|
histone cluster 1, H4k
|
|
chrX_+_34971918
|
0.351
|
NM_201236
|
Rhox4e
|
reproductive homeobox 4E
|
|
chr5_-_130363128
|
0.332
|
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2
|
|
chr11_-_106642357
|
0.322
|
|
Ddx5
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
|
|
chr9_+_77602337
|
0.312
|
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit
|
|
chr8_+_113618575
|
0.255
|
NM_198308
|
Pdpr
|
pyruvate dehydrogenase phosphatase regulatory subunit
|
|
chrX_+_34805369
|
0.223
|
NM_001039688
|
Rhox4a
|
reproductive homeobox 4A
|
|
chr16_-_21995441
|
0.214
|
NM_153404
NM_001083894
|
Liph
|
lipase, member H
|
|
chr11_-_11798035
|
0.205
|
|
Ddc
|
dopa decarboxylase
|
|
chr12_-_111934448
|
0.193
|
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1
|
|
chr9_+_43118845
|
0.162
|
NM_023655
|
Trim29
|
tripartite motif-containing 29
|
|
chrX_+_34847000
|
0.141
|
NM_021300
|
Rhox4e
Rhox4b
Rhox4d
|
reproductive homeobox 4E
reproductive homeobox 4B
reproductive homeobox 4D
|
|
chrX_-_151524332
|
0.138
|
NM_020015
|
Magea1
|
melanoma antigen, family A, 1
|
|
chr10_+_80102813
|
0.109
|
|
Csnk1g2
|
casein kinase 1, gamma 2
|
|
chr15_+_57747523
|
0.108
|
|
Wdr67
|
WD repeat domain 67
|
|
chr2_+_29933228
|
0.106
|
|
Pkn3
|
protein kinase N3
|
|
chr7_-_5068497
|
0.099
|
NM_001145013
NM_138954
|
Rfpl4
|
ret finger protein-like 4
|
|
chr15_-_96886327
|
0.096
|
NM_027052
|
Slc38a4
|
solute carrier family 38, member 4
|
|
chr14_-_53110720
|
0.080
|
NM_020512
|
Olfr1507
|
olfactory receptor 1507
|
|
chr19_-_42358648
|
0.075
|
|
Crtac1
|
cartilage acidic protein 1
|
|
chr5_-_33560819
|
0.043
|
NM_133903
|
Spon2
|
spondin 2, extracellular matrix protein
|
|
chr19_+_11611327
|
0.020
|
NM_025658
|
Ms4a4d
|
membrane-spanning 4-domains, subfamily A, member 4D
|
|
chr10_-_90586710
|
0.019
|
|
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3
|
|
chr2_-_48804672
|
0.019
|
NM_001177313
NM_011958
|
Orc4
|
origin recognition complex, subunit 4
|