|
chr2_+_172978566
|
19.947
|
NM_011044
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic
|
|
chr7_+_50699457
|
7.881
|
NM_026695
|
Etfb
|
electron transferring flavoprotein, beta polypeptide
|
|
chr11_+_70471297
|
7.504
|
NM_007933
|
Eno3
|
enolase 3, beta muscle
|
|
chr9_+_108564418
|
7.477
|
NM_020520
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20
|
|
chr15_-_27560282
|
7.367
|
NM_001013792
|
Fam105b
|
family with sequence similarity 105, member B
|
|
chr11_+_74462965
|
6.683
|
NM_001081158
|
1300001I01Rik
|
RIKEN cDNA 1300001I01 gene
|
|
chr18_+_70728508
|
6.648
|
|
Mbd2
|
methyl-CpG binding domain protein 2
|
|
chr11_+_70471347
|
5.870
|
|
Eno3
|
enolase 3, beta muscle
|
|
chr16_+_48872720
|
5.730
|
NM_181596
|
Retnlg
|
resistin like gamma
|
|
chr2_+_92215590
|
4.892
|
|
Pex16
|
peroxisomal biogenesis factor 16
|
|
chr2_-_6466116
|
4.861
|
|
Celf2
|
CUGBP, Elav-like family member 2
|
|
chr14_+_122933538
|
4.715
|
NM_144844
|
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide
|
|
chr14_+_122933560
|
4.707
|
|
Pcca
|
propionyl-Coenzyme A carboxylase, alpha polypeptide
|
|
chr4_+_59594569
|
4.620
|
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2
|
|
chr16_+_44139908
|
4.563
|
NM_028108
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit
|
|
chr7_+_31017305
|
4.420
|
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I
|
|
chr2_+_92215395
|
4.346
|
NM_145122
|
Pex16
|
peroxisomal biogenesis factor 16
|
|
chr17_-_34164996
|
4.251
|
NM_013543
|
H2-Ke6
|
H2-K region expressed gene 6
|
|
chr18_+_74938827
|
4.124
|
NM_177470
|
Acaa2
|
acetyl-Coenzyme A acyltransferase 2 (mitochondrial 3-oxoacyl-Coenzyme A thiolase)
|
|
chr6_+_70557402
|
4.053
|
|
|
|
|
chr5_-_31999977
|
4.010
|
NM_153196
|
Rbks
|
ribokinase
|
|
chr19_-_6996189
|
3.884
|
NM_007953
|
Esrra
|
estrogen related receptor, alpha
|
|
chr15_-_77363703
|
3.877
|
NM_175391
|
Apol7c
|
apolipoprotein L 7c
|
|
chr2_+_157854457
|
3.557
|
NM_027434
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr3_+_115591053
|
3.488
|
NM_027193
|
Dph5
|
DPH5 homolog (S. cerevisiae)
|
|
chr3_-_8923789
|
3.391
|
NM_025434
|
Mrps28
|
mitochondrial ribosomal protein S28
|
|
chr3_-_58329780
|
3.353
|
NM_030685
|
Serp1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr4_-_11003327
|
3.348
|
NM_001085493
|
2310030N02Rik
|
RIKEN cDNA 2310030N02 gene
|
|
chr4_+_122960059
|
3.167
|
NM_130881
|
Pabpc4
|
poly(A) binding protein, cytoplasmic 4
|
|
chr17_+_87762817
|
3.097
|
|
Ttc7
|
tetratricopeptide repeat domain 7
|
|
chr4_+_97444326
|
3.052
|
|
Nfia
|
nuclear factor I/A
|
|
chr5_+_135663405
|
3.002
|
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr7_-_25873560
|
2.991
|
|
|
|
|
chr4_+_118102305
|
2.897
|
NM_001039176
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr15_-_89259756
|
2.861
|
|
Chkb
|
choline kinase beta
|
|
chr7_+_31017092
|
2.850
|
NM_027259
|
Polr2i
|
polymerase (RNA) II (DNA directed) polypeptide I
|
|
chr19_+_10116104
|
2.841
|
|
Fads3
|
fatty acid desaturase 3
|
|
chr19_+_10116079
|
2.838
|
|
Fads3
|
fatty acid desaturase 3
|
|
chr5_+_58109278
|
2.832
|
|
Pcdh7
|
protocadherin 7
|
|
chr19_-_3912772
|
2.826
|
|
Ndufs8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8
|
|
chr6_+_124662387
|
2.764
|
|
Phb2
|
prohibitin 2
|
|
chr10_+_13272699
|
2.758
|
NM_025748
|
Adat2
|
adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae)
|
|
chr7_+_100023762
|
2.750
|
NM_028243
|
Prcp
|
prolylcarboxypeptidase (angiotensinase C)
|
|
chr1_+_74552600
|
2.739
|
NM_021383
|
Rqcd1
|
rcd1 (required for cell differentiation) homolog 1 (S. pombe)
|
|
chr3_+_116216008
|
2.709
|
|
Dbt
|
dihydrolipoamide branched chain transacylase E2
|
|
chr19_-_3912727
|
2.706
|
|
Ndufs8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8
|
|
chr9_-_46043339
|
2.608
|
NM_023114
|
Apoc3
|
apolipoprotein C-III
|
|
chr1_+_55145132
|
2.600
|
|
Hspe1
|
heat shock protein 1 (chaperonin 10)
|
|
chr1_-_55144687
|
2.597
|
NM_010477
|
Hspd1
|
heat shock protein 1 (chaperonin)
|
|
chr15_-_102019373
|
2.582
|
|
Csad
|
cysteine sulfinic acid decarboxylase
|
|
chr9_+_20807180
|
2.550
|
NM_023167
|
Mrpl4
|
mitochondrial ribosomal protein L4
|
|
chr19_+_38470458
|
2.533
|
NM_175507
|
Tmem20
|
transmembrane protein 20
|
|
chr1_+_177562307
|
2.532
|
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr10_-_13272899
|
2.528
|
NM_001164195
NM_019961
|
Pex3
|
peroxisomal biogenesis factor 3
|
|
chr4_+_97444563
|
2.471
|
|
Nfia
|
nuclear factor I/A
|
|
chr19_-_3912710
|
2.452
|
NM_144870
|
Ndufs8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8
|
|
chr16_+_48872756
|
2.428
|
|
Retnlg
|
resistin like gamma
|
|
chr11_-_83406475
|
2.423
|
NM_009139
|
Ccl6
|
chemokine (C-C motif) ligand 6
|
|
chr3_+_116215963
|
2.423
|
NM_010022
|
Dbt
|
dihydrolipoamide branched chain transacylase E2
|
|
chr1_+_169548381
|
2.411
|
NM_001159731
|
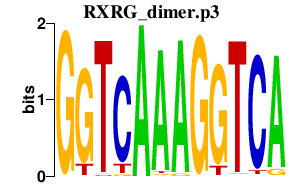
Rxrg
|
retinoid X receptor gamma
|
|
chr9_-_44226119
|
2.386
|
|
Ccdc84
|
coiled-coil domain containing 84
|
|
chr5_+_135663096
|
2.383
|
NM_011714
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B
|
|
chr10_-_93052348
|
2.370
|
NM_027246
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F
|
|
chr9_+_123429291
|
2.323
|
|
Limd1
|
LIM domains containing 1
|
|
chr2_-_145760400
|
2.317
|
NM_025820
|
Crnkl1
|
Crn, crooked neck-like 1 (Drosophila)
|
|
chr2_+_153171730
|
2.308
|
NM_001039939
|
Asxl1
|
additional sex combs like 1 (Drosophila)
|
|
chr9_-_21940154
|
2.264
|
NM_001102405
|
Acp5
|
acid phosphatase 5, tartrate resistant
|
|
chr9_+_118415358
|
2.259
|
NM_018748
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4
|
|
chr1_-_55144637
|
2.249
|
|
Hspd1
|
heat shock protein 1 (chaperonin)
|
|
chr4_+_59594516
|
2.237
|
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2
|
|
chr13_-_73465894
|
2.216
|
NM_010888
|
Ndufs6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6
|
|
chr4_-_131628011
|
2.203
|
NM_183428
|
Epb4.1
|
erythrocyte protein band 4.1
|
|
chrX_-_39265047
|
2.191
|
NM_001033422
|
Thoc2
|
THO complex 2
|
|
chr15_-_100253413
|
2.168
|
NM_001146161
NM_008732
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2
|
|
chr2_-_164630371
|
2.165
|
NM_133240
|
Acot8
|
acyl-CoA thioesterase 8
|
|
chr7_+_53152702
|
2.113
|
|
Tmem143
|
transmembrane protein 143
|
|
chr10_-_13272820
|
2.084
|
|
Pex3
|
peroxisomal biogenesis factor 3
|
|
chr11_+_98245124
|
2.074
|
NM_011540
|
Tcap
|
titin-cap
|
|
chr9_-_44226073
|
2.071
|
NM_201372
|
Ccdc84
|
coiled-coil domain containing 84
|
|
chr5_+_36807807
|
2.016
|
NM_024478
|
Grpel1
|
GrpE-like 1, mitochondrial
|
|
chrX_+_98644459
|
2.013
|
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr5_-_134652558
|
2.001
|
NM_033572
|
Wbscr16
|
Williams-Beuren syndrome chromosome region 16 homolog (human)
|
|
chr12_-_105990160
|
1.995
|
NM_148948
|
Dicer1
|
Dicer1, Dcr-1 homolog (Drosophila)
|
|
chr18_+_78546492
|
1.934
|
|
Gm6133
|
60S ribosomal protein L17 pseudogene
|
|
chr8_-_88079180
|
1.888
|
|
Dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr16_+_4684069
|
1.886
|
NM_001135112
NM_023646
|
Dnaja3
|
DnaJ (Hsp40) homolog, subfamily A, member 3
|
|
chr6_+_85381958
|
1.878
|
NM_144918
|
Smyd5
|
SET and MYND domain containing 5
|
|
chr13_+_49282844
|
1.869
|
NM_013610
|
Ninj1
|
ninjurin 1
|
|
chr8_-_88079141
|
1.869
|
|
Dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr3_-_115591014
|
1.851
|
|
A930005H10Rik
|
RIKEN cDNA A930005H10 gene
|
|
chr1_+_57463391
|
1.822
|
NM_001081181
|
9430016H08Rik
|
RIKEN cDNA 9430016H08 gene
|
|
chr3_-_58329639
|
1.812
|
|
Serp1
|
stress-associated endoplasmic reticulum protein 1
|
|
chr4_+_97444450
|
1.806
|
|
Nfia
|
nuclear factor I/A
|
|
chr1_+_177562321
|
1.800
|
NM_133809
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase)
|
|
chr2_+_75670242
|
1.773
|
|
Agps
|
alkylglycerone phosphate synthase
|
|
chr2_-_164630347
|
1.771
|
|
Acot8
|
acyl-CoA thioesterase 8
|
|
chr10_-_119549945
|
1.762
|
NM_080446
|
Helb
|
helicase (DNA) B
|
|
chr5_+_32000312
|
1.746
|
NM_144541
NM_181279
NM_181280
NM_181281
NM_181282
|
Bre
|
brain and reproductive organ-expressed protein
|
|
chr8_-_88079169
|
1.700
|
NM_019794
|
Dnaja2
|
DnaJ (Hsp40) homolog, subfamily A, member 2
|
|
chr4_+_97444312
|
1.693
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr11_+_105043195
|
1.684
|
|
Tlk2
|
tousled-like kinase 2 (Arabidopsis)
|
|
chr9_+_20807312
|
1.678
|
|
Mrpl4
|
mitochondrial ribosomal protein L4
|
|
chr10_+_84380305
|
1.637
|
NM_183172
|
Ric8b
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr2_+_30122256
|
1.623
|
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr14_-_65515555
|
1.563
|
|
Hmbox1
|
homeobox containing 1
|
|
chr7_-_97089575
|
1.559
|
NM_026304
|
l7Rn6
|
lethal, Chr 7, Rinchik 6
|
|
chr5_+_32794685
|
1.552
|
|
Ppp1cb
|
protein phosphatase 1, catalytic subunit, beta isoform
|
|
chr8_+_13026014
|
1.524
|
NM_010172
|
F7
|
coagulation factor VII
|
|
chr3_-_115590869
|
1.487
|
|
A930005H10Rik
|
RIKEN cDNA A930005H10 gene
|
|
chr2_+_30122046
|
1.479
|
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr1_+_74552794
|
1.464
|
|
Rqcd1
|
rcd1 (required for cell differentiation) homolog 1 (S. pombe)
|
|
chr3_+_122598292
|
1.458
|
NM_007980
|
Fabp2
|
fatty acid binding protein 2, intestinal
|
|
chr4_+_59594461
|
1.457
|
|
Hsdl2
|
hydroxysteroid dehydrogenase like 2
|
|
chr2_-_73749080
|
1.444
|
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9)
|
|
chr10_-_117282901
|
1.444
|
|
Rap1b
|
RAS related protein 1b
|
|
chr1_-_140744146
|
1.432
|
NM_010714
|
Lhx9
|
LIM homeobox protein 9
|
|
chr10_+_84380371
|
1.416
|
NM_001013441
|
Ric8b
|
resistance to inhibitors of cholinesterase 8 homolog B (C. elegans)
|
|
chr15_-_102019443
|
1.412
|
NM_144942
|
Csad
|
cysteine sulfinic acid decarboxylase
|
|
chr11_+_101303558
|
1.410
|
NM_011289
|
Rpl27
|
ribosomal protein L27
|
|
chr4_-_136604609
|
1.374
|
NM_198248
|
Zbtb40
|
zinc finger and BTB domain containing 40
|
|
chr4_+_151307819
|
1.330
|
NM_172704
|
Dnajc11
|
DnaJ (Hsp40) homolog, subfamily C, member 11
|
|
chr10_+_127799210
|
1.318
|
|
Cs
|
citrate synthase
|
|
chr15_-_75839859
|
1.307
|
NM_001168253
|
Fam83h
|
family with sequence similarity 83, member H
|
|
chr15_-_82160509
|
1.301
|
|
Naga
|
N-acetyl galactosaminidase, alpha
|
|
chr15_-_42508294
|
1.299
|
NM_009640
|
Angpt1
|
angiopoietin 1
|
|
chr2_+_75670230
|
1.290
|
NM_172666
|
Agps
|
alkylglycerone phosphate synthase
|
|
chr15_+_97622089
|
1.255
|
|
Slc48a1
|
solute carrier family 48 (heme transporter), member 1
|
|
chr1_+_55144949
|
1.252
|
NM_008303
|
Hspe1
|
heat shock protein 1 (chaperonin 10)
|
|
chr11_-_4604308
|
1.219
|
NM_197979
|
Uqcr10
|
ubiquinol-cytochrome c reductase, complex III subunit X
|
|
chr10_+_45009110
|
1.188
|
NM_024286
|
Popdc3
|
popeye domain containing 3
|
|
chr2_-_73749317
|
1.180
|
NM_175015
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9)
|
|
chr12_+_21322219
|
1.169
|
NM_026347
|
Iah1
|
isoamyl acetate-hydrolyzing esterase 1 homolog (S. cerevisiae)
|
|
chr11_-_69828831
|
1.156
|
NM_017366
|
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain
|
|
chr14_-_52634206
|
1.136
|
|
Zfp219
|
zinc finger protein 219
|
|
chr10_-_14425271
|
1.121
|
NM_025418
|
Vta1
|
Vps20-associated 1 homolog (S. cerevisiae)
|
|
chr4_-_147818818
|
1.118
|
NM_027873
|
Ubiad1
|
UbiA prenyltransferase domain containing 1
|
|
chr12_+_9581302
|
1.094
|
|
Osr1
|
odd-skipped related 1 (Drosophila)
|
|
chr18_-_36904173
|
1.070
|
|
Ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2
|
|
chr18_-_36904178
|
1.060
|
NM_010885
|
Ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2
|
|
chr12_+_85379311
|
1.048
|
NM_134247
|
Acot4
|
acyl-CoA thioesterase 4
|
|
chr7_+_38968235
|
1.046
|
NM_001085385
|
1600014C10Rik
|
RIKEN cDNA 1600014C10 gene
|
|
chr13_+_43466084
|
1.024
|
NM_178848
|
Sirt5
|
sirtuin 5 (silent mating type information regulation 2 homolog) 5 (S. cerevisiae)
|
|
chr1_+_136090015
|
1.022
|
NM_016749
|
Mybph
|
myosin binding protein H
|
|
chr1_+_193645351
|
1.020
|
NM_010892
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr8_-_90483392
|
1.011
|
NM_033327
|
Zfp423
|
zinc finger protein 423
|
|
chr9_-_70782614
|
0.979
|
NM_008280
|
Lipc
|
lipase, hepatic
|
|
chr4_+_141302635
|
0.974
|
|
Agmat
|
agmatine ureohydrolase (agmatinase)
|
|
chr2_-_164568585
|
0.973
|
NM_027961
|
Wfdc3
|
WAP four-disulfide core domain 3
|
|
chr11_+_21138879
|
0.958
|
NM_139061
|
Vps54
|
vacuolar protein sorting 54 (yeast)
|
|
chr2_+_157854427
|
0.908
|
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr2_-_24775083
|
0.906
|
NM_001012518
NM_001109686
NM_001109687
NM_172545
|
Ehmt1
|
euchromatic histone methyltransferase 1
|
|
chr1_-_63223345
|
0.899
|
NM_001160038
NM_001160039
NM_001160040
NM_145518
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1
|
|
chr2_-_157854168
|
0.878
|
|
Tti1
|
Tel2 interacting protein 1 homolog (S. pombe)
|
|
chr15_-_81702298
|
0.871
|
NM_026737
|
Phf5a
|
PHD finger protein 5A
|
|
chr8_+_131238027
|
0.865
|
|
Itgb1
|
integrin beta 1 (fibronectin receptor beta)
|
|
chr4_+_97444494
|
0.850
|
|
Nfia
|
nuclear factor I/A
|
|
chr7_-_20535921
|
0.836
|
NM_027308
|
2210010C17Rik
|
RIKEN cDNA 2210010C17 gene
|
|
chr9_-_71959599
|
0.832
|
NM_011544
|
Tcf12
|
transcription factor 12
|
|
chr5_+_117570113
|
0.830
|
|
Taok3
|
TAO kinase 3
|
|
chr3_+_120994646
|
0.821
|
NM_024178
|
Alg14
|
asparagine-linked glycosylation 14 homolog (yeast)
|
|
chr2_+_30122064
|
0.798
|
NM_172267
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1
|
|
chr11_+_32646291
|
0.780
|
|
Fbxw11
|
F-box and WD-40 domain protein 11
|
|
chr3_-_144595486
|
0.776
|
NM_001033199
|
AI747448
|
expressed sequence AI747448
|
|
chr8_+_13037791
|
0.775
|
|
F10
|
coagulation factor X
|
|
chr6_+_83958578
|
0.760
|
NM_001077694
|
Dysf
|
dysferlin
|
|
chr3_+_58329742
|
0.752
|
NM_001005509
|
Eif2a
|
eukaryotic translation initiation factor 2A
|
|
chr3_+_132754791
|
0.750
|
NM_027927
|
Ints12
|
integrator complex subunit 12
|
|
chr4_+_118081908
|
0.742
|
NM_020000
NM_173719
|
Med8
|
mediator of RNA polymerase II transcription, subunit 8 homolog (yeast)
|
|
chr15_-_57907967
|
0.740
|
NM_001042438
NM_009572
|
Zhx1
|
zinc fingers and homeoboxes 1
|
|
chr2_+_35122282
|
0.733
|
|
Gsn
|
gelsolin
|
|
chr3_-_88138354
|
0.726
|
NM_024246
|
Tmem79
|
transmembrane protein 79
|
|
chr6_-_91423388
|
0.698
|
NM_133928
|
Chchd4
|
coiled-coil-helix-coiled-coil-helix domain containing 4
|
|
chr19_+_29441927
|
0.696
|
NM_021893
|
Cd274
|
CD274 antigen
|
|
chr7_-_20300595
|
0.674
|
|
Tomm40
|
translocase of outer mitochondrial membrane 40 homolog (yeast)
|
|
chr1_+_173319048
|
0.659
|
NM_001005508
|
Arhgap30
|
Rho GTPase activating protein 30
|
|
chr9_-_70782366
|
0.655
|
|
Lipc
|
lipase, hepatic
|
|
chr19_-_32177613
|
0.642
|
NM_018830
|
Asah2
|
N-acylsphingosine amidohydrolase 2
|
|
chr10_+_78037170
|
0.625
|
NM_173751
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like
|
|
chr4_+_118032592
|
0.617
|
|
Hyi
|
hydroxypyruvate isomerase homolog (E. coli)
|
|
chr11_-_78235686
|
0.613
|
NM_022411
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2
|
|
chr2_+_35139423
|
0.607
|
|
Gsn
|
gelsolin
|
|
chr9_-_86589782
|
0.596
|
NM_008615
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr11_+_86358491
|
0.592
|
NM_001199045
NM_001199046
NM_001199047
NM_019756
|
Tubd1
|
tubulin, delta 1
|
|
chr7_-_111507159
|
0.584
|
NM_175648
|
Trim30b
|
tripartite motif-containing 30B
|
|
chr4_-_94316806
|
0.583
|
NM_175305
|
Lrrc19
|
leucine rich repeat containing 19
|
|
chr3_+_96633987
|
0.573
|
NM_021517
|
Pdzk1
|
PDZ domain containing 1
|
|
chr14_+_55059393
|
0.555
|
|
Mmp14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr2_-_103637714
|
0.554
|
|
Caprin1
|
cell cycle associated protein 1
|
|
chr2_+_35139479
|
0.534
|
|
Gsn
|
gelsolin
|
|
chr4_-_57155985
|
0.531
|
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr9_-_44575861
|
0.502
|
NM_145985
|
Arcn1
|
archain 1
|
|
chr11_+_57917712
|
0.502
|
|
Cnot8
|
CCR4-NOT transcription complex, subunit 8
|
|
chr1_-_63223190
|
0.489
|
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1
|
|
chrX_+_12869324
|
0.489
|
|
Ddx3x
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 3, X-linked
|
|
chr4_-_40883509
|
0.483
|
|
Bag1
|
BCL2-associated athanogene 1
|
|
chr6_-_42323208
|
0.459
|
NM_023580
|
Epha1
|
Eph receptor A1
|
|
chr2_+_157854434
|
0.455
|
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B
|
|
chr4_+_19502296
|
0.443
|
|
Fam82b
|
family with sequence similarity 82, member B
|
|
chr11_+_84684395
|
0.438
|
|
Gm10374
|
predicted gene 10374
|
|
chr2_+_18594027
|
0.438
|
NM_147778
|
Commd3
|
COMM domain containing 3
|