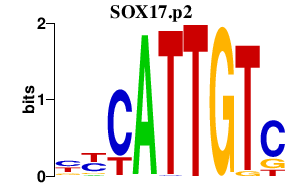
|
chr19_+_11482121
|
22.996
|
NM_029499
|
Ms4a4c
|
membrane-spanning 4-domains, subfamily A, member 4C
|
|
chr4_+_134024234
|
20.988
|
NM_019641
|
Stmn1
|
stathmin 1
|
|
chr17_-_51878720
|
18.853
|
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr7_-_52622323
|
16.901
|
|
|
|
|
chr16_-_17125198
|
15.123
|
NM_183287
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene
|
|
chr16_+_52031704
|
14.738
|
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr9_-_70269246
|
14.389
|
|
Ccnb2
|
cyclin B2
|
|
chr11_+_68859125
|
13.779
|
NM_011496
|
Aurkb
|
aurora kinase B
|
|
chr7_-_80703275
|
13.592
|
|
1810026B05Rik
|
RIKEN cDNA 1810026B05 gene
|
|
chr1_+_34516650
|
13.443
|
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18
|
|
chr6_+_124780226
|
12.964
|
|
Cdca3
|
cell division cycle associated 3
|
|
chr1_+_34516590
|
12.873
|
NM_011206
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18
|
|
chr13_-_101556356
|
12.673
|
NM_172301
|
Ccnb1
|
cyclin B1
|
|
chr6_+_124780193
|
12.057
|
NM_013538
|
Cdca3
|
cell division cycle associated 3
|
|
chr8_-_13677450
|
11.776
|
NM_009025
|
Rasa3
|
RAS p21 protein activator 3
|
|
chr11_+_23156022
|
11.581
|
NM_001035226
NM_134014
|
Xpo1
|
exportin 1, CRM1 homolog (yeast)
|
|
chr3_-_107135939
|
11.246
|
NM_001045807
|
Rbm15
|
RNA binding motif protein 15
|
|
chr11_-_116274264
|
11.060
|
NM_027258
|
Rnf157
|
ring finger protein 157
|
|
chr11_+_11586215
|
11.049
|
NM_001025597
NM_009578
|
Ikzf1
|
IKAROS family zinc finger 1
|
|
chr4_-_151851624
|
10.832
|
|
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chr4_-_133443662
|
10.794
|
NM_009097
|
Rps6ka1
|
ribosomal protein S6 kinase polypeptide 1
|
|
chr11_-_106021399
|
10.772
|
NM_172397
|
Limd2
|
LIM domain containing 2
|
|
chr9_+_83728331
|
10.393
|
|
Ttk
|
Ttk protein kinase
|
|
chr7_-_133848235
|
10.342
|
NM_009898
|
Coro1a
|
coronin, actin binding protein 1A
|
|
chr5_+_30969259
|
10.097
|
NM_007681
|
Cenpa
|
centromere protein A
|
|
chr11_-_98190774
|
10.080
|
NM_010895
|
Neurod2
|
neurogenic differentiation 2
|
|
chr16_+_52031661
|
9.975
|
NM_001033238
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr3_+_96025040
|
9.938
|
NM_178214
|
Hist2h2be
|
histone cluster 2, H2be
|
|
chr9_+_83728295
|
9.887
|
NM_001110265
NM_009445
|
Ttk
|
Ttk protein kinase
|
|
chr6_+_29644178
|
9.829
|
NM_146173
|
Tspan33
|
tetraspanin 33
|
|
chr11_-_104330747
|
9.524
|
|
1700081L11Rik
|
RIKEN cDNA 1700081L11 gene
|
|
chr17_-_25023253
|
9.133
|
NM_011956
|
Nubp2
|
nucleotide binding protein 2
|
|
chr11_+_98769122
|
9.115
|
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr7_-_90883999
|
9.030
|
NM_010551
|
Il16
|
interleukin 16
|
|
chr4_-_124614072
|
9.005
|
NM_026560
|
Cdca8
|
cell division cycle associated 8
|
|
chr6_-_86619001
|
8.972
|
NM_010751
|
Mxd1
|
MAX dimerization protein 1
|
|
chr8_-_107813357
|
8.805
|
NM_001166394
NM_028888
|
4931428F04Rik
|
RIKEN cDNA 4931428F04 gene
|
|
chr7_+_13466525
|
8.788
|
NM_153802
|
Zfp128
|
zinc finger protein 128
|
|
chr2_+_144649401
|
8.746
|
NM_025490
|
1700010M22Rik
|
RIKEN cDNA 1700010M22 gene
|
|
chr14_+_56236989
|
8.714
|
NM_020002
|
Rec8
|
REC8 homolog (yeast)
|
|
chr14_-_47809063
|
8.681
|
NM_008102
|
Gch1
|
GTP cyclohydrolase 1
|
|
chr12_-_56087322
|
8.557
|
NM_013815
|
Baz1a
|
bromodomain adjacent to zinc finger domain 1A
|
|
chr7_+_149135990
|
8.554
|
NM_001009929
NM_001009930
|
Brsk2
|
BR serine/threonine kinase 2
|
|
chr9_+_72655019
|
8.443
|
NM_175485
|
Prtg
|
protogenin homolog (Gallus gallus)
|
|
chr7_+_3332984
|
8.383
|
|
Cacng7
|
calcium channel, voltage-dependent, gamma subunit 7
|
|
chr3_+_89904315
|
8.064
|
|
|
|
|
chr2_+_146047656
|
8.017
|
NM_016889
|
Insm1
|
insulinoma-associated 1
|
|
chr9_-_70269326
|
7.898
|
NM_007630
|
Ccnb2
|
cyclin B2
|
|
chr14_+_55634290
|
7.880
|
NM_026890
|
Ngdn
|
neuroguidin, EIF4E binding protein
|
|
chr11_-_116274124
|
7.853
|
|
Rnf157
|
ring finger protein 157
|
|
chr7_-_25654709
|
7.838
|
NM_182785
|
Lypd4
|
Ly6/Plaur domain containing 4
|
|
chr11_+_98769162
|
7.837
|
NM_001025779
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr6_+_145070255
|
7.735
|
NM_008511
|
Lrmp
|
lymphoid-restricted membrane protein
|
|
chr4_+_134676559
|
7.712
|
NM_019732
|
Runx3
|
runt related transcription factor 3
|
|
chr19_-_15999377
|
7.591
|
NM_177420
|
Psat1
|
phosphoserine aminotransferase 1
|
|
chr19_+_37509290
|
7.565
|
NM_008245
|
Hhex
|
hematopoietically expressed homeobox
|
|
chr1_-_185857753
|
7.448
|
NM_028516
|
1700056E22Rik
|
RIKEN cDNA 1700056E22 gene
|
|
chr5_-_8622882
|
7.426
|
NM_198620
|
Rundc3b
|
RUN domain containing 3B
|
|
chr10_+_111497244
|
7.192
|
NM_027018
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1
|
|
chr1_+_19093102
|
7.087
|
NM_153154
|
Tcfap2d
|
transcription factor AP-2, delta
|
|
chr11_-_114657186
|
7.046
|
NM_028055
|
Btbd17
|
BTB (POZ) domain containing 17
|
|
chr3_-_68848657
|
7.031
|
NM_025863
|
Trim59
|
tripartite motif-containing 59
|
|
chr17_-_24854956
|
6.939
|
NM_026205
|
Rnf151
|
ring finger protein 151
|
|
chr3_-_36470904
|
6.871
|
NM_009828
|
Ccna2
|
cyclin A2
|
|
chr11_-_98885428
|
6.858
|
NM_011623
|
Top2a
|
topoisomerase (DNA) II alpha
|
|
chr18_-_6516073
|
6.847
|
NM_007935
|
Epc1
|
enhancer of polycomb homolog 1 (Drosophila)
|
|
chr3_-_68848588
|
6.817
|
|
Trim59
|
tripartite motif-containing 59
|
|
chr17_+_87762817
|
6.799
|
|
Ttc7
|
tetratricopeptide repeat domain 7
|
|
chr2_-_39046224
|
6.798
|
NM_178778
|
Scai
|
suppressor of cancer cell invasion
|
|
chrX_+_90900201
|
6.761
|
NM_177546
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform
|
|
chrX_-_7151340
|
6.754
|
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr7_-_139508762
|
6.605
|
NM_029935
|
Chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr2_+_155890933
|
6.600
|
NM_139151
|
Spag4
|
sperm associated antigen 4
|
|
chr11_+_98769464
|
6.576
|
NM_011799
|
Cdc6
|
cell division cycle 6 homolog (S. cerevisiae)
|
|
chr2_+_3630729
|
6.571
|
NM_025626
|
Fam107b
|
family with sequence similarity 107, member B
|
|
chr10_+_53316720
|
6.559
|
NM_025541
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr3_-_36470845
|
6.530
|
|
Ccna2
|
cyclin A2
|
|
chr17_-_25864310
|
6.496
|
NM_145409
|
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 homolog (S. cerevisiae)
|
|
chr16_+_91225794
|
6.493
|
NM_016967
|
Olig2
|
oligodendrocyte transcription factor 2
|
|
chr2_-_5635547
|
6.416
|
NM_177343
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr3_-_36470882
|
6.409
|
|
Ccna2
|
cyclin A2
|
|
chr3_+_9250566
|
6.362
|
NM_177660
|
Zbtb10
|
zinc finger and BTB domain containing 10
|
|
chr16_+_44765848
|
6.345
|
NM_021325
|
Cd200r1
|
CD200 receptor 1
|
|
chr1_-_167932735
|
6.295
|
NM_011137
NM_198932
|
Pou2f1
|
POU domain, class 2, transcription factor 1
|
|
chr2_+_112124767
|
6.274
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr4_+_154335029
|
6.271
|
NM_010419
|
Hes5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr3_-_105970438
|
6.236
|
NM_009892
|
Chi3l3
|
chitinase 3-like 3
|
|
chr7_+_89319717
|
6.231
|
NM_017400
|
Sh3gl3
|
SH3-domain GRB2-like 3
|
|
chr7_+_17460697
|
6.222
|
|
Dact3
|
dapper homolog 3, antagonist of beta-catenin (xenopus)
|
|
chr9_-_62644026
|
6.203
|
|
Fem1b
|
feminization 1 homolog b (C. elegans)
|
|
chr7_-_56892061
|
6.126
|
NM_001005232
|
Dbx1
|
developing brain homeobox 1
|
|
chr6_-_148894953
|
6.121
|
NM_019643
|
Fam60a
|
family with sequence similarity 60, member A
|
|
chr12_+_25393083
|
6.045
|
NM_009104
|
Rrm2
|
ribonucleotide reductase M2
|
|
chr9_+_13632152
|
6.030
|
NM_176836
|
Fam76b
|
family with sequence similarity 76, member B
|
|
chr9_+_65738150
|
5.902
|
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene
|
|
chr10_-_94407211
|
5.872
|
NM_018797
|
Plxnc1
|
plexin C1
|
|
chr10_+_84218792
|
5.865
|
NM_001024918
|
Rfx4
|
regulatory factor X, 4 (influences HLA class II expression)
|
|
chr5_-_138721164
|
5.848
|
NM_172412
|
Gpc2
|
glypican 2 (cerebroglycan)
|
|
chr9_-_103382586
|
5.845
|
NM_001002896
|
Bfsp2
|
beaded filament structural protein 2, phakinin
|
|
chr6_+_125095236
|
5.829
|
NM_178787
NM_001039669
|
Iffo1
|
intermediate filament family orphan 1
|
|
chr12_-_76697166
|
5.819
|
NM_012024
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B (B56), epsilon isoform
|
|
chr4_-_118109775
|
5.817
|
NM_023223
|
Cdc20
|
cell division cycle 20 homolog (S. cerevisiae)
|
|
chrX_+_89735287
|
5.766
|
NM_001034103
|
Gm5941
|
predicted gene 5941
|
|
chr13_-_56236756
|
5.758
|
|
H2afy
|
H2A histone family, member Y
|
|
chr2_+_167340686
|
5.723
|
|
Rnf114
|
ring finger protein 114
|
|
chr2_+_91366966
|
5.722
|
|
Ckap5
|
cytoskeleton associated protein 5
|
|
chr3_-_83844042
|
5.720
|
NM_172681
|
D930015E06Rik
|
RIKEN cDNA D930015E06 gene
|
|
chr2_-_5635517
|
5.720
|
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr17_-_53828596
|
5.698
|
NM_028232
|
Sgol1
|
shugoshin-like 1 (S. pombe)
|
|
chr17_-_37712365
|
5.649
|
NM_146289
|
Olfr113
|
olfactory receptor 113
|
|
chr10_+_90039435
|
5.638
|
NM_001177396
NM_001177398
NM_181398
|
Ipw
Anks1b
|
imprinted gene in the Prader-Willi syndrome region
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr11_-_49526211
|
5.626
|
NM_212484
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6
|
|
chr9_-_83334633
|
5.622
|
NM_027448
NM_029434
|
Lca5
|
Leber congenital amaurosis 5 (human)
|
|
chr11_-_102786433
|
5.621
|
NM_197959
|
Kif18b
|
kinesin family member 18B
|
|
chr4_-_151851574
|
5.617
|
NM_010598
|
Kcnab2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2
|
|
chrX_-_7151185
|
5.613
|
NM_138605
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F
|
|
chr3_+_89586320
|
5.600
|
|
Ube2q1
|
ubiquitin-conjugating enzyme E2Q (putative) 1
|
|
chr10_+_40069113
|
5.598
|
NM_001168304
NM_198164
|
Cdk19
|
cyclin-dependent kinase 19
|
|
chr11_-_102786403
|
5.559
|
|
Kif18b
|
kinesin family member 18B
|
|
chr2_+_150735306
|
5.551
|
NM_001163476
NM_027014
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog)
|
|
chr10_-_80962400
|
5.516
|
|
S1pr4
|
sphingosine-1-phosphate receptor 4
|
|
chr1_-_36502079
|
5.515
|
NM_001013374
|
Lman2l
|
lectin, mannose-binding 2-like
|
|
chr7_+_148033703
|
5.507
|
NM_027019
|
Odf3
|
outer dense fiber of sperm tails 3
|
|
chr9_+_27204405
|
5.483
|
NM_029299
|
Spata19
|
spermatogenesis associated 19
|
|
chr4_+_11118500
|
5.475
|
NM_001037134
|
Ccne2
|
cyclin E2
|
|
chr7_-_140793797
|
5.458
|
NM_028654
|
4930404H21Rik
|
RIKEN cDNA 4930404H21 gene
|
|
chr2_+_118872854
|
5.435
|
NM_029617
|
Casc5
|
cancer susceptibility candidate 5
|
|
chr19_+_11518042
|
5.423
|
NM_021718
|
Ms4a4b
|
membrane-spanning 4-domains, subfamily A, member 4B
|
|
chrX_+_72341192
|
5.422
|
NM_001030307
|
Dkc1
|
dyskeratosis congenita 1, dyskerin homolog (human)
|
|
chr4_+_108132677
|
5.403
|
|
Zcchc11
|
zinc finger, CCHC domain containing 11
|
|
chr9_-_21564742
|
5.402
|
|
Spc24
|
SPC24, NDC80 kinetochore complex component, homolog (S. cerevisiae)
|
|
chr7_-_31319977
|
5.373
|
NM_178252
|
Arhgap33
|
Rho GTPase activating protein 33
|
|
chr16_-_46120221
|
5.347
|
NM_032465
|
Cd96
|
CD96 antigen
|
|
chr9_+_118387306
|
5.323
|
NM_001164789
NM_010136
|
Eomes
|
eomesodermin homolog (Xenopus laevis)
|
|
chr7_-_139508857
|
5.296
|
|
Chst15
|
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15
|
|
chr2_-_5635228
|
5.241
|
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr17_-_35975148
|
5.234
|
NM_011655
|
Tubb5
|
tubulin, beta 5
|
|
chr11_+_86922732
|
5.225
|
NM_025377
|
Fam33a
|
family with sequence similarity 33, member A
|
|
chr2_+_4322020
|
5.205
|
NM_001177843
|
Frmd4a
|
FERM domain containing 4A
|
|
chr4_-_22415277
|
5.203
|
NM_008899
|
Pou3f2
|
POU domain, class 3, transcription factor 2
|
|
chr11_-_86922163
|
5.182
|
NM_175563
|
Prr11
|
proline rich 11
|
|
chr17_-_24607134
|
5.149
|
|
E4f1
4833447I15Rik
|
E4F transcription factor 1
RIKEN cDNA 4833447I15 gene
|
|
chr3_+_94964364
|
5.132
|
|
Sema6c
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6C
|
|
chr9_+_65738168
|
5.122
|
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene
|
|
chr5_-_124170947
|
5.111
|
|
Zcchc8
|
zinc finger, CCHC domain containing 8
|
|
chr8_-_83262881
|
5.103
|
|
Smarca5
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5
|
|
chr9_-_123883223
|
5.098
|
|
Ccr1
|
chemokine (C-C motif) receptor 1
|
|
chr17_-_79754427
|
5.093
|
NM_026514
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr5_+_114581164
|
5.078
|
NM_011677
|
Ung
|
uracil DNA glycosylase
|
|
chr13_-_56236910
|
5.060
|
NM_001159513
NM_001159514
NM_001159515
NM_012015
|
H2afy
|
H2A histone family, member Y
|
|
chr6_-_83267540
|
5.043
|
NM_008638
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase
|
|
chr11_-_75161767
|
5.035
|
NM_001164223
NM_026653
|
Rpa1
|
replication protein A1
|
|
chr10_-_80005514
|
5.025
|
|
Rexo1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae)
|
|
chr17_-_24388262
|
5.015
|
NM_007634
|
Ccnf
|
cyclin F
|
|
chr5_-_23536487
|
4.996
|
NM_053090
|
Fam126a
|
family with sequence similarity 126, member A
|
|
chr7_-_52622373
|
4.991
|
NM_029741
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr17_-_79754229
|
4.981
|
|
Cdc42ep3
|
CDC42 effector protein (Rho GTPase binding) 3
|
|
chr8_+_77633426
|
4.958
|
NM_008566
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae)
|
|
chr11_+_94828911
|
4.939
|
NM_018792
|
Hils1
|
histone H1-like protein in spermatids 1
|
|
chr7_-_15208414
|
4.933
|
NM_001039146
|
Vmn1r90
|
vomeronasal 1 receptor 90
|
|
chr5_+_28492235
|
4.932
|
NM_010134
|
En2
|
engrailed 2
|
|
chr10_+_96079634
|
4.924
|
NM_007569
|
Btg1
|
B-cell translocation gene 1, anti-proliferative
|
|
chr12_-_76697148
|
4.921
|
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B (B56), epsilon isoform
|
|
chr17_+_36002537
|
4.904
|
NM_001146711
|
2310014H01Rik
|
RIKEN cDNA 2310014H01 gene
|
|
chr9_-_123883398
|
4.884
|
NM_009912
|
Ccr1
|
chemokine (C-C motif) receptor 1
|
|
chr15_-_79572885
|
4.867
|
NM_194342
|
Sun2
|
Sad1 and UNC84 domain containing 2
|
|
chr7_+_109614267
|
4.816
|
|
Rrm1
|
ribonucleotide reductase M1
|
|
chr9_+_78243583
|
4.815
|
NM_001191044
|
Ddx43
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 43
|
|
chr2_+_158492468
|
4.803
|
NM_153089
|
Ppp1r16b
|
protein phosphatase 1, regulatory (inhibitor) subunit 16B
|
|
chr13_-_97440734
|
4.792
|
NM_008255
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase
|
|
chr7_-_52494453
|
4.786
|
|
Cd37
|
CD37 antigen
|
|
chr3_+_137527448
|
4.784
|
NM_016750
|
H2afz
|
H2A histone family, member Z
|
|
chr9_-_32348863
|
4.773
|
NM_008026
|
Fli1
|
Friend leukemia integration 1
|
|
chr5_+_114580397
|
4.768
|
NM_001040691
|
Ung
|
uracil DNA glycosylase
|
|
chr12_-_115286643
|
4.759
|
|
Igh-V11
|
immunoglobulin heavy chain (V11 family)
|
|
chr19_+_41904459
|
4.739
|
NM_008043
|
Frat1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr3_-_9833502
|
4.731
|
NM_001195031
NM_053182
|
Pag1
|
phosphoprotein associated with glycosphingolipid microdomains 1
|
|
chr3_+_89904553
|
4.700
|
|
Tpm3
|
tropomyosin 3, gamma
|
|
chr5_-_123350242
|
4.698
|
NM_013910
|
Kdm2b
|
lysine (K)-specific demethylase 2B
|
|
chr4_+_109650629
|
4.692
|
NM_172296
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2
|
|
chr4_+_132686434
|
4.671
|
NM_153423
|
Wasf2
|
WAS protein family, member 2
|
|
chr6_+_139990045
|
4.665
|
NM_007605
|
Capza3
|
capping protein (actin filament) muscle Z-line, alpha 3
|
|
chr4_-_154417257
|
4.663
|
|
Plch2
|
phospholipase C, eta 2
|
|
chr10_+_43198945
|
4.651
|
NM_199028
|
Bend3
|
BEN domain containing 3
|
|
chr2_-_72931338
|
4.647
|
|
|
|
|
chr4_-_115796249
|
4.636
|
NM_001122958
NM_009015
NM_001122959
|
Rad54l
|
RAD54 like (S. cerevisiae)
|
|
chr2_+_92024338
|
4.633
|
NM_138755
|
Phf21a
|
PHD finger protein 21A
|
|
chr6_+_86354212
|
4.631
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr17_-_33897428
|
4.624
|
NM_008997
|
Rab11b
|
RAB11B, member RAS oncogene family
|
|
chr12_-_76697214
|
4.592
|
|
Ppp2r5e
|
protein phosphatase 2, regulatory subunit B (B56), epsilon isoform
|
|
chr1_+_193645351
|
4.588
|
NM_010892
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2
|
|
chr5_-_100467945
|
4.549
|
|
Hnrpdl
|
heterogeneous nuclear ribonucleoprotein D-like
|
|
chr11_-_119948078
|
4.537
|
|
Azi1
|
5-azacytidine induced gene 1
|
|
chr13_-_41165827
|
4.536
|
NM_008547
|
Mak
|
male germ cell-associated kinase
|
|
chr4_-_154041819
|
4.533
|
NM_028513
|
Actrt2
|
actin-related protein T2
|
|
chr7_+_97357406
|
4.520
|
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein
|
|
chr7_-_52622289
|
4.519
|
|
Ppfia3
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 3
|
|
chr16_-_50591201
|
4.505
|
NM_027046
|
Ccdc54
|
coiled-coil domain containing 54
|
|
chr10_-_122550401
|
4.493
|
|
Usp15
|
ubiquitin specific peptidase 15
|
|
chr4_+_120204835
|
4.492
|
NM_177570
|
Slfnl1
|
schlafen like 1
|