|
chr9_+_120058842
|
48.487
|
NM_001039364
NM_001039365
NM_008614
|
Mobp
|
myelin-associated oligodendrocytic basic protein
|
|
chr17_-_51878720
|
30.302
|
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr12_-_73337534
|
27.861
|
|
Rtn1
|
reticulon 1
|
|
chr18_-_43219062
|
23.566
|
NM_028392
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr18_+_37157533
|
20.873
|
NM_138661
|
Pcdha9
|
protocadherin alpha 9
|
|
chrX_+_155897136
|
20.371
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chrX_+_132424640
|
19.898
|
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B9
|
|
chr17_-_90436488
|
19.059
|
|
Nrxn1
|
neurexin I
|
|
chr2_-_170232008
|
17.517
|
|
Bcas1
|
breast carcinoma amplified sequence 1
|
|
chr18_-_23197156
|
17.084
|
NM_001161483
|
Nol4
|
nucleolar protein 4
|
|
chrX_+_155897056
|
16.717
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chrX_-_149078963
|
16.214
|
|
Shroom2
|
shroom family member 2
|
|
chr18_+_37120093
|
16.075
|
NM_009959
|
Pcdha5
|
protocadherin alpha 5
|
|
chr12_+_91570569
|
15.454
|
|
Nrxn3
|
neurexin III
|
|
chr5_+_67067477
|
14.810
|
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr18_+_82723920
|
14.755
|
|
Mbp
|
myelin basic protein
|
|
chr9_-_49311823
|
14.704
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr18_+_37112301
|
14.460
|
NM_001174154
NM_007766
|
Pcdha4-g
Pcdha4
|
protocadherin alpha 4-gamma
protocadherin alpha 4
|
|
chr10_-_30375572
|
14.288
|
NM_001111267
|
Ncoa7
|
nuclear receptor coactivator 7
|
|
chr18_+_37127284
|
14.093
|
NM_007767
|
Pcdha6
|
protocadherin alpha 6
|
|
chr5_+_89012596
|
13.419
|
NM_027530
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr10_+_20760625
|
13.273
|
NM_001177776
|
Ahi1
|
Abelson helper integration site 1
|
|
chr9_+_109905895
|
12.984
|
|
|
|
|
chr10_+_119256444
|
12.879
|
NM_028736
NM_130891
|
Grip1
|
glutamate receptor interacting protein 1
|
|
chr18_+_37133455
|
12.873
|
NM_009957
|
Pcdha7
|
protocadherin alpha 7
|
|
chr10_+_28863801
|
12.810
|
NM_026138
|
6330407J23Rik
|
RIKEN cDNA 6330407J23 gene
|
|
chr11_+_87574041
|
12.576
|
NM_172449
|
Bzrap1
|
benzodiazepine receptor associated protein 1
|
|
chr15_+_98465328
|
12.159
|
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr5_+_19413335
|
12.156
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr2_-_165709697
|
12.114
|
|
Zmynd8
|
zinc finger, MYND-type containing 8
|
|
chrX_+_162782484
|
11.868
|
NM_001177955
NM_001177956
NM_001177957
NM_001177958
NM_001177959
NM_001177960
|
Gpm6b
|
glycoprotein m6b
|
|
chr12_-_73337707
|
11.717
|
NM_001007596
|
Rtn1
|
reticulon 1
|
|
chr5_+_37637027
|
11.667
|
NM_007765
|
Crmp1
|
collapsin response mediator protein 1
|
|
chr5_+_67067497
|
11.557
|
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr11_-_97372344
|
11.406
|
|
Srcin1
|
SRC kinase signaling inhibitor 1
|
|
chr7_-_38554642
|
11.364
|
NM_172385
|
Zfp536
|
zinc finger protein 536
|
|
chrX_+_130829929
|
11.226
|
|
Tmem35
|
transmembrane protein 35
|
|
chr4_-_91042997
|
11.184
|
NM_001177883
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr5_+_27231234
|
11.066
|
NM_001198886
|
Dpp6
|
dipeptidylpeptidase 6
|
|
chr18_+_37644674
|
11.046
|
NM_053142
|
Pcdhb17
|
protocadherin beta 17
|
|
chr18_+_37098858
|
11.040
|
NM_198117
|
Pcdha2
|
protocadherin alpha 2
|
|
chr1_-_42749940
|
10.876
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr8_+_125935470
|
10.769
|
|
Tubb3
|
tubulin, beta 3
|
|
chr10_-_70055559
|
10.736
|
NM_001162846
|
Phyhipl
|
phytanoyl-CoA hydroxylase interacting protein-like
|
|
chr16_-_18622083
|
10.721
|
|
|
|
|
chr18_+_37152024
|
10.559
|
NM_201243
|
Pcdha8
|
protocadherin alpha 8
|
|
chr18_+_82723854
|
10.556
|
NM_001025251
NM_001025255
NM_001025256
NM_001025258
NM_001025259
NM_001025254
|
Mbp
|
myelin basic protein
|
|
chr1_+_160292470
|
10.554
|
NM_007495
|
Astn1
|
astrotactin 1
|
|
chr15_+_98465193
|
10.469
|
NM_007581
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr18_+_82723907
|
10.361
|
|
Mbp
|
myelin basic protein
|
|
chr12_+_30213224
|
10.239
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr5_+_89012547
|
10.206
|
|
Rufy3
|
RUN and FYVE domain containing 3
|
|
chr4_-_109960078
|
10.139
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr2_+_143371807
|
10.131
|
NM_008792
|
Pcsk2
|
proprotein convertase subtilisin/kexin type 2
|
|
chrX_+_130829763
|
10.019
|
NM_026239
|
Tmem35
|
transmembrane protein 35
|
|
chr9_-_75531734
|
9.970
|
NM_001164790
NM_009130
|
Scg3
|
secretogranin III
|
|
chr16_-_46120221
|
9.934
|
NM_032465
|
Cd96
|
CD96 antigen
|
|
chr3_+_8509479
|
9.835
|
NM_025285
|
Stmn2
|
stathmin-like 2
|
|
chr14_+_60997104
|
9.765
|
NM_001164705
NM_028113
|
Fam123a
|
family with sequence similarity 123, member A
|
|
chr5_-_23012407
|
9.633
|
|
Srpk2
|
serine/arginine-rich protein specific kinase 2
|
|
chr13_-_111070225
|
9.554
|
|
Rab3c
|
RAB3C, member RAS oncogene family
|
|
chr6_+_54222814
|
9.434
|
NM_023543
|
Chn2
|
chimerin (chimaerin) 2
|
|
chr7_-_82453166
|
9.324
|
|
Sv2b
|
synaptic vesicle glycoprotein 2 b
|
|
chr19_-_42160732
|
9.316
|
NM_198108
|
Morn4
|
MORN repeat containing 4
|
|
chr8_+_65430028
|
9.242
|
NM_023689
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3
|
|
chr5_+_112684838
|
9.239
|
NM_023695
|
Crybb1
|
crystallin, beta B1
|
|
chr2_+_14310024
|
9.211
|
NM_001012305
|
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12
|
|
chr7_+_89319681
|
9.188
|
|
Sh3gl3
|
SH3-domain GRB2-like 3
|
|
chr9_-_21820860
|
9.143
|
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr10_+_61245998
|
9.124
|
|
|
|
|
chr7_+_89319717
|
9.069
|
NM_017400
|
Sh3gl3
|
SH3-domain GRB2-like 3
|
|
chr1_+_160292665
|
9.065
|
|
Astn1
|
astrotactin 1
|
|
chr3_-_80606500
|
8.909
|
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chr11_-_77538789
|
8.848
|
NM_009965
|
Cryba1
|
crystallin, beta A1
|
|
chr9_+_46721685
|
8.799
|
|
2900052N01Rik
|
RIKEN cDNA 2900052N01 gene
|
|
chr12_+_30219769
|
8.662
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr12_+_3365295
|
8.639
|
|
Kif3c
|
kinesin family member 3C
|
|
chr14_-_9142289
|
8.611
|
NM_183187
|
Fam107a
|
family with sequence similarity 107, member A
|
|
chr14_+_60997156
|
8.524
|
|
Fam123a
|
family with sequence similarity 123, member A
|
|
chr10_-_115910911
|
8.480
|
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr4_-_91066674
|
8.436
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr15_-_88808967
|
8.368
|
NM_133241
|
Mlc1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 homolog (human)
|
|
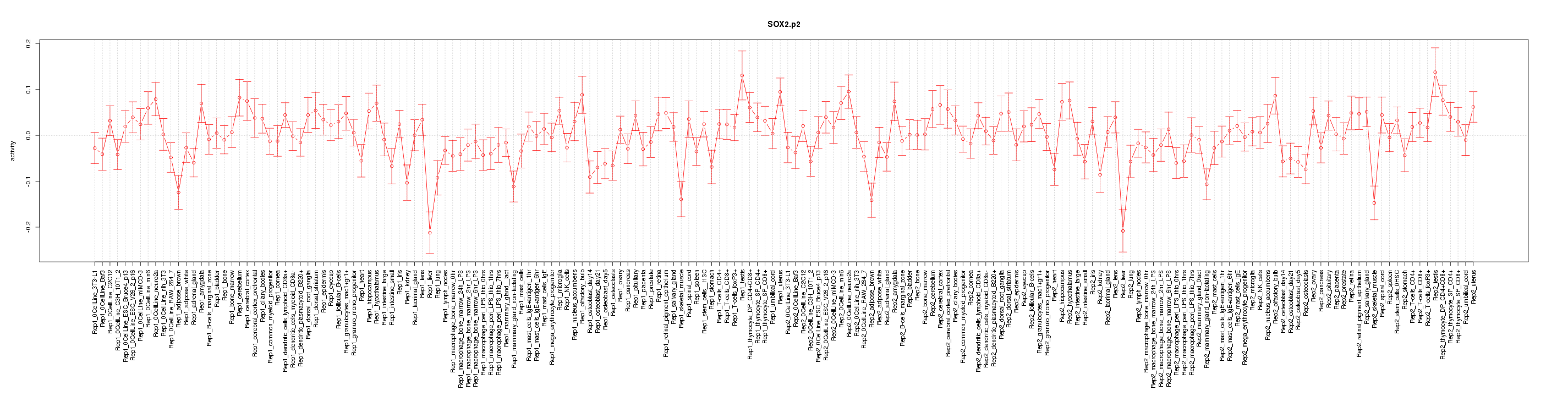
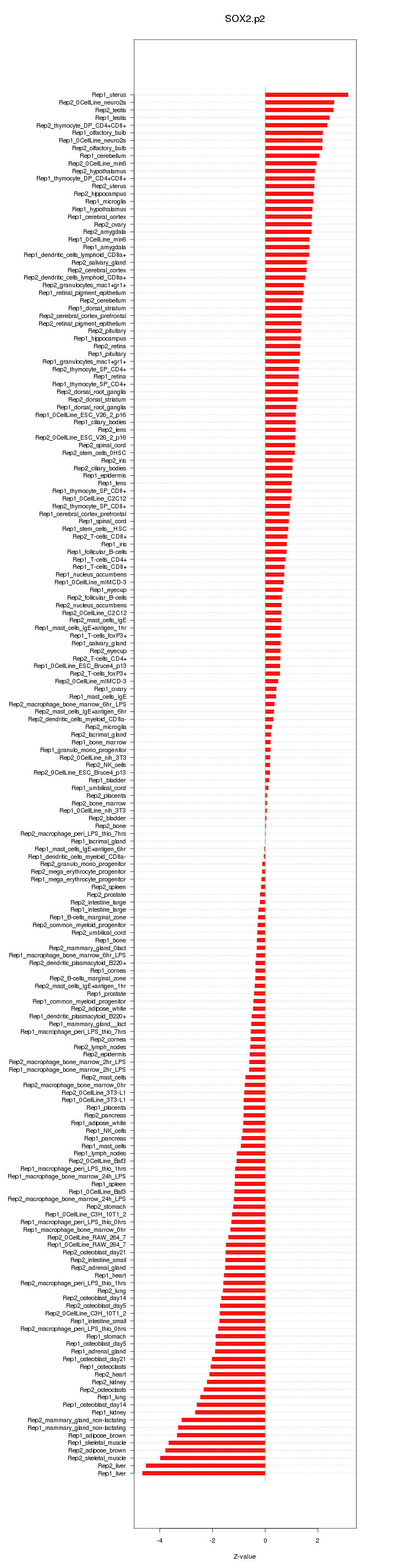
chr3_+_34548916
|
8.339
|
NM_011443
|
Sox2
|
SRY-box containing gene 2
|
|
chr12_+_3365147
|
8.338
|
|
Kif3c
|
kinesin family member 3C
|
|
chr5_+_67067359
|
8.283
|
NM_011670
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr9_-_52484889
|
8.216
|
|
AI593442
|
expressed sequence AI593442
|
|
chr14_-_122097583
|
8.212
|
NM_001081039
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr6_-_18464737
|
8.135
|
|
Cttnbp2
|
cortactin binding protein 2
|
|
chr9_-_29219306
|
8.114
|
|
Ntm
|
neurotrimin
|
|
chrX_+_10173557
|
8.114
|
|
|
|
|
chr6_+_86354212
|
8.099
|
NM_001164078
NM_001164079
NM_011585
|
Tia1
|
cytotoxic granule-associated RNA binding protein 1
|
|
chr9_-_49607164
|
8.029
|
NM_001081445
NM_001113204
NM_010875
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr9_-_75458865
|
7.955
|
|
Tmod2
|
tropomodulin 2
|
|
chr8_+_56040553
|
7.932
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr14_-_80171003
|
7.892
|
NM_001042726
NM_021543
|
Pcdh8
|
protocadherin 8
|
|
chr8_+_10153910
|
7.775
|
NM_001081397
|
Myo16
|
myosin XVI
|
|
chr18_-_77870871
|
7.756
|
|
2310007O11Rik
|
RIKEN cDNA 2310007O11 gene
|
|
chr1_+_129702056
|
7.742
|
|
Ccnt2
|
cyclin T2
|
|
chr10_-_115910578
|
7.737
|
NM_021452
|
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4
|
|
chr6_-_135664978
|
7.696
|
|
|
|
|
chr2_+_179777363
|
7.672
|
|
Ss18l1
|
synovial sarcoma translocation gene on chromosome 18-like 1
|
|
chr7_-_107475377
|
7.633
|
|
|
|
|
chr12_-_36768992
|
7.618
|
|
Tspan13
|
tetraspanin 13
|
|
chr9_-_60497537
|
7.611
|
|
Lrrc49
|
leucine rich repeat containing 49
|
|
chrX_+_139952975
|
7.599
|
NM_001195048
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr7_-_89079168
|
7.587
|
|
Hdgfrp3
|
hepatoma-derived growth factor, related protein 3
|
|
chr9_-_29219382
|
7.577
|
|
Ntm
|
neurotrimin
|
|
chr9_+_111277323
|
7.573
|
|
Trank1
|
tetratricopeptide repeat and ankyrin repeat containing 1
|
|
chr18_-_43847367
|
7.510
|
NM_001163637
|
Jakmip2
|
janus kinase and microtubule interacting protein 2
|
|
chr17_-_51951378
|
7.477
|
NM_001163630
|
Satb1
|
special AT-rich sequence binding protein 1
|
|
chr4_-_151235861
|
7.440
|
NM_001081557
NM_001195565
|
Camta1
|
calmodulin binding transcription activator 1
|
|
chr18_+_59222034
|
7.419
|
NM_001167925
NM_173759
|
A730017C20Rik
|
RIKEN cDNA A730017C20 gene
|
|
chr4_+_43521765
|
7.414
|
|
Car9
|
carbonic anhydrase 9
|
|
chr14_-_20796199
|
7.412
|
|
Gng2
|
guanine nucleotide binding protein (G protein), gamma 2
|
|
chr1_+_17135421
|
7.397
|
NM_010267
|
Gdap1
|
ganglioside-induced differentiation-associated-protein 1
|
|
chr12_-_36769054
|
7.331
|
NM_025359
|
Tspan13
|
tetraspanin 13
|
|
chr3_+_13471681
|
7.311
|
|
Ralyl
|
RALY RNA binding protein-like
|
|
chr8_+_98226914
|
7.305
|
NM_001195006
NM_145602
|
Ndrg4
|
N-myc downstream regulated gene 4
|
|
chr1_-_75250520
|
7.268
|
|
Ptprn
|
protein tyrosine phosphatase, receptor type, N
|
|
chr2_+_121134843
|
7.214
|
|
Mtap1a
|
microtubule-associated protein 1 A
|
|
chr7_+_97535516
|
7.164
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr18_-_43552694
|
7.152
|
NM_009468
|
Dpysl3
|
dihydropyrimidinase-like 3
|
|
chr8_+_56040445
|
7.084
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr7_-_89079335
|
7.004
|
NM_013886
|
Hdgfrp3
|
hepatoma-derived growth factor, related protein 3
|
|
chr2_-_65405546
|
6.932
|
NM_018732
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha
|
|
chr14_+_84933363
|
6.918
|
|
Pcdh17
|
protocadherin 17
|
|
chr14_-_109313333
|
6.857
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr9_+_107948476
|
6.829
|
|
Ip6k1
|
inositol hexaphosphate kinase 1
|
|
chr14_-_109313298
|
6.824
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr9_-_49607017
|
6.718
|
|
Ncam1
|
neural cell adhesion molecule 1
|
|
chr7_+_69493148
|
6.667
|
NM_010882
|
Ndn
|
necdin
|
|
chr7_+_98239427
|
6.633
|
|
Dlg2
|
discs, large homolog 2 (Drosophila)
|
|
chr14_-_49282546
|
6.574
|
NM_144841
|
Otx2
|
orthodenticle homolog 2 (Drosophila)
|
|
chrX_+_90900201
|
6.514
|
NM_177546
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform
|
|
chr8_-_87461251
|
6.511
|
NM_019945
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr6_+_143156520
|
6.488
|
|
Etnk1
|
ethanolamine kinase 1
|
|
chr4_+_109650629
|
6.481
|
NM_172296
|
Dmrta2
|
doublesex and mab-3 related transcription factor like family A2
|
|
chr15_-_98783909
|
6.470
|
NM_011653
|
Tuba1a
|
tubulin, alpha 1A
|
|
chr10_-_115922360
|
6.402
|
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2
|
|
chr9_-_21822333
|
6.397
|
|
Elavl3
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 3 (Hu antigen C)
|
|
chr11_-_79317471
|
6.389
|
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr3_+_105776628
|
6.377
|
NM_007696
|
Ovgp1
|
oviductal glycoprotein 1
|
|
chr7_+_97535490
|
6.318
|
NM_001040087
NM_001040088
|
Sytl2
|
synaptotagmin-like 2
|
|
chr15_+_91958558
|
6.311
|
NM_007727
|
Cntn1
|
contactin 1
|
|
chr1_-_195196423
|
6.292
|
NM_144817
|
Camk1g
|
calcium/calmodulin-dependent protein kinase I gamma
|
|
chr18_+_37901716
|
6.282
|
NM_033578
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6
|
|
chr15_+_4325490
|
6.271
|
NM_177355
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3
|
|
chr6_+_141472906
|
6.249
|
NM_001177772
NM_021471
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1
|
|
chr11_+_54128404
|
6.236
|
NM_001033599
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6
|
|
chr7_+_90015842
|
6.208
|
NM_175366
|
Mex3b
|
mex3 homolog B (C. elegans)
|
|
chr2_+_157385835
|
6.183
|
NM_010923
NM_180960
|
Nnat
|
neuronatin
|
|
chr18_+_37089837
|
6.179
|
NM_054072
|
Pcdha1
|
protocadherin alpha 1
|
|
chr10_+_90292388
|
6.173
|
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr1_-_175297624
|
6.165
|
|
Cadm3
|
cell adhesion molecule 3
|
|
chr5_+_72091188
|
6.137
|
NM_008069
|
Gabrb1
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 1
|
|
chr10_+_69388242
|
6.135
|
NM_009670
NM_170687
|
Ank3
|
ankyrin 3, epithelial
|
|
chr4_-_120781830
|
6.126
|
NM_001081312
|
Tmco2
|
transmembrane and coiled-coil domains 2
|
|
chr1_+_40082201
|
6.113
|
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_-_34926055
|
6.101
|
|
Uba2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr4_+_129190877
|
6.098
|
|
Marcksl1
|
MARCKS-like 1
|
|
chr10_+_53316720
|
6.071
|
NM_025541
|
Asf1a
|
ASF1 anti-silencing function 1 homolog A (S. cerevisiae)
|
|
chr1_-_175297778
|
6.065
|
NM_053199
|
Cadm3
|
cell adhesion molecule 3
|
|
chr4_+_126850123
|
6.019
|
|
Dlgap3
|
discs, large (Drosophila) homolog-associated protein 3
|
|
chr16_+_7409879
|
5.988
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr7_+_107423813
|
5.981
|
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr6_+_107479719
|
5.973
|
NM_008516
|
Lrrn1
|
leucine rich repeat protein 1, neuronal
|
|
chr19_+_38339167
|
5.972
|
NM_020278
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr16_+_13986691
|
5.941
|
NM_144518
|
2900011O08Rik
|
RIKEN cDNA 2900011O08 gene
|
|
chr7_+_114514258
|
5.929
|
NM_021889
|
Syt9
|
synaptotagmin IX
|
|
chr17_+_70871444
|
5.915
|
NM_001128180
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr17_-_35975075
|
5.886
|
|
Tubb5
|
tubulin, beta 5
|
|
chr10_+_89336253
|
5.880
|
NM_001128086
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr9_-_103664100
|
5.860
|
NM_178638
|
Tmem108
|
transmembrane protein 108
|
|
chr3_-_80606641
|
5.818
|
|
Gria2
|
glutamate receptor, ionotropic, AMPA2 (alpha 2)
|
|
chrX_+_133527693
|
5.795
|
NM_001035509
NM_001035510
NM_025893
|
Zcchc18
|
zinc finger, CCHC domain containing 18
|
|
chr18_+_37644390
|
5.794
|
|
Pcdhb17
|
protocadherin beta 17
|
|
chr4_+_129190872
|
5.791
|
|
Marcksl1
|
MARCKS-like 1
|
|
chr6_+_30758582
|
5.771
|
|
Copg2as2
|
coatomer protein complex, subunit gamma 2, antisense 2
|
|
chr9_-_75458954
|
5.760
|
|
Tmod2
|
tropomodulin 2
|
|
chr8_-_87461220
|
5.755
|
|
Mast1
|
microtubule associated serine/threonine kinase 1
|
|
chr11_+_3230733
|
5.750
|
NM_178149
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1
|
|
chr6_-_12699189
|
5.749
|
NM_001164805
|
Thsd7a
|
thrombospondin, type I, domain containing 7A
|
|
chr10_-_80693488
|
5.728
|
NM_178662
|
Atcay
|
ataxia, cerebellar, Cayman type homolog (human)
|
|
chr18_+_35750071
|
5.711
|
|
Matr3
|
matrin 3
|
|
chr11_-_79317583
|
5.704
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr7_-_34926107
|
5.694
|
|
Uba2
|
ubiquitin-like modifier activating enzyme 2
|
|
chr4_+_98213324
|
5.684
|
NM_001005787
NM_007704
|
Inadl
|
InaD-like (Drosophila)
|
|
chr15_+_99532787
|
5.683
|
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1
|
|
chr1_+_9591550
|
5.654
|
|
3110035E14Rik
|
RIKEN cDNA 3110035E14 gene
|
|
chr2_+_112124767
|
5.649
|
NM_133648
|
Slc12a6
|
solute carrier family 12, member 6
|
|
chr5_+_135526788
|
5.638
|
|
|
|
|
chr6_-_128531620
|
5.632
|
NM_001001179
|
BC048546
|
cDNA sequence BC048546
|
|
chr1_-_42724504
|
5.599
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr9_-_108363229
|
5.594
|
|
Klhdc8b
|
kelch domain containing 8B
|
|
chr19_+_22767272
|
5.585
|
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3
|
|
chr12_+_25516433
|
5.570
|
NM_001083341
NM_026037
|
Mboat2
|
membrane bound O-acyltransferase domain containing 2
|
|
chr7_+_25266986
|
5.564
|
NM_153112
|
Cadm4
|
cell adhesion molecule 4
|
|
chr11_-_69635657
|
5.561
|
NM_028855
|
Spem1
|
sperm maturation 1
|
|
chr13_-_111070344
|
5.541
|
|
Rab3c
|
RAB3C, member RAS oncogene family
|
|
chr3_-_95032039
|
5.470
|
|
Mllt11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|