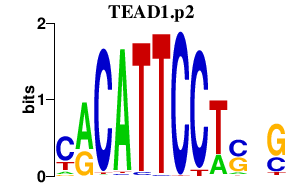
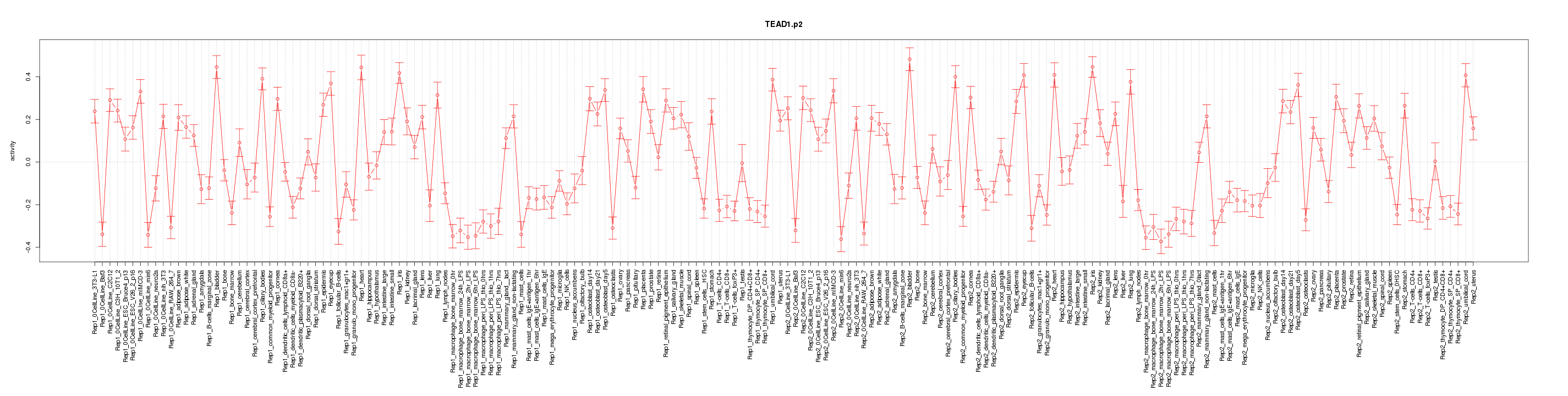
|
chr10_+_24315247
|
246.453
|
NM_010217
|
Ctgf
|
connective tissue growth factor
|
|
chr19_-_34329804
|
149.834
|
NM_007392
|
Acta2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr7_-_34918076
|
110.877
|
NM_207212
|
Wtip
|
WT1-interacting protein
|
|
chr9_+_102620058
|
98.651
|
|
Amotl2
|
angiomotin-like 2
|
|
chr9_+_102620051
|
96.685
|
|
Amotl2
|
angiomotin-like 2
|
|
chr9_+_102620096
|
93.251
|
NM_019764
|
Amotl2
|
angiomotin-like 2
|
|
chr11_-_55233354
|
90.836
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr2_+_156601175
|
87.579
|
NM_172118
|
Myl9
|
myosin, light polypeptide 9, regulatory
|
|
chr2_+_156601150
|
86.792
|
|
Myl9
|
myosin, light polypeptide 9, regulatory
|
|
chr1_-_45559904
|
82.987
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_+_51346086
|
74.629
|
|
Sdpr
|
serum deprivation response
|
|
chr6_+_17256383
|
74.130
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr6_+_17256369
|
73.479
|
NM_007616
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr1_+_51345952
|
72.733
|
NM_138741
|
Sdpr
|
serum deprivation response
|
|
chr9_-_66896961
|
66.899
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr11_-_55233581
|
66.759
|
NM_009242
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr7_+_137789668
|
66.588
|
|
Tacc2
|
transforming, acidic coiled-coil containing protein 2
|
|
chr11_-_32122236
|
64.618
|
|
Rhbdf1
|
rhomboid family 1 (Drosophila)
|
|
chr11_-_32122259
|
63.525
|
NM_010117
|
Rhbdf1
|
rhomboid family 1 (Drosophila)
|
|
chr11_-_55233366
|
63.387
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr6_+_34548512
|
60.860
|
|
Cald1
|
caldesmon 1
|
|
chr8_-_49075884
|
59.825
|
NM_133791
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2
|
|
chr1_-_45559866
|
57.911
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr3_-_103541868
|
57.294
|
|
Olfml3
|
olfactomedin-like 3
|
|
chr3_-_145312928
|
56.135
|
NM_010516
|
Cyr61
|
cysteine rich protein 61
|
|
chr7_-_116760069
|
55.809
|
|
St5
|
suppression of tumorigenicity 5
|
|
chr1_+_51346108
|
55.678
|
|
Sdpr
|
serum deprivation response
|
|
chr11_-_100831909
|
54.569
|
NM_008986
|
Ptrf
|
polymerase I and transcript release factor
|
|
chr1_-_45559700
|
53.823
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr7_-_116760660
|
53.356
|
NM_001001326
NM_029811
|
St5
|
suppression of tumorigenicity 5
|
|
chr8_+_95376459
|
53.195
|
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr3_-_103541882
|
52.633
|
NM_133859
|
Olfml3
|
olfactomedin-like 3
|
|
chr11_-_55233339
|
51.254
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr16_+_5007426
|
49.861
|
|
Gm5480
|
predicted gene 5480
|
|
chr6_+_17256415
|
49.425
|
|
Cav1
|
caveolin 1, caveolae protein
|
|
chr1_-_45560001
|
48.014
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr2_-_127482430
|
47.901
|
NM_001171187
NM_010762
|
Mal
|
myelin and lymphocyte protein, T-cell differentiation protein
|
|
chr6_+_88674821
|
45.238
|
|
Mgll
|
monoglyceride lipase
|
|
chrX_-_73120485
|
44.780
|
NM_001166453
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr17_-_35838818
|
42.335
|
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr9_-_66896992
|
42.256
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr11_+_62944973
|
41.894
|
NM_008885
|
Pmp22
|
peripheral myelin protein 22
|
|
chr8_-_64238843
|
41.659
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr7_-_91733736
|
41.505
|
|
Fah
|
fumarylacetoacetate hydrolase
|
|
chr3_-_151929210
|
41.440
|
NM_199465
|
Nexn
|
nexilin
|
|
chr1_+_137732910
|
41.257
|
|
Tnnt2
|
troponin T2, cardiac
|
|
chr1_+_12682510
|
41.201
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr9_-_66897015
|
41.144
|
NM_001164248
NM_001164249
NM_001164250
NM_001164251
NM_001164255
NM_024427
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr15_+_78673024
|
40.060
|
NM_027219
|
Cdc42ep1
|
CDC42 effector protein (Rho GTPase binding) 1
|
|
chr7_-_31839825
|
39.828
|
NM_052991
NM_194321
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1
|
|
chrX_-_73120447
|
39.316
|
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr14_+_31692725
|
38.311
|
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr6_+_122463595
|
37.192
|
|
Mfap5
|
microfibrillar associated protein 5
|
|
chr1_-_170362298
|
37.027
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr5_+_136363769
|
36.905
|
NM_013560
|
Hspb1
|
heat shock protein 1
|
|
chr1_+_45368382
|
35.462
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr5_-_107718503
|
34.068
|
NM_011578
|
Tgfbr3
|
transforming growth factor, beta receptor III
|
|
chr11_+_87394609
|
33.530
|
NM_011129
|
Sept4
|
septin 4
|
|
chr1_+_195128163
|
33.263
|
NM_008484
|
Lamb3
|
laminin, beta 3
|
|
chr17_+_47573557
|
33.250
|
NM_145489
|
AI661453
|
expressed sequence AI661453
|
|
chr6_+_90424727
|
32.903
|
|
Klf15
|
Kruppel-like factor 15
|
|
chr9_-_8004559
|
32.670
|
NM_001171147
NM_009534
|
Yap1
|
yes-associated protein 1
|
|
chr19_-_4439350
|
32.645
|
NM_007485
|
Rhod
|
ras homolog gene family, member D
|
|
chr7_-_133606617
|
32.538
|
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr14_+_31692419
|
32.081
|
NM_181390
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1
|
|
chr14_+_47502637
|
32.021
|
NM_001037221
NM_028966
|
Samd4
|
sterile alpha motif domain containing 4
|
|
chr3_+_68272097
|
31.902
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chr11_+_87394543
|
31.310
|
|
Sept4
|
septin 4
|
|
chr9_-_58884114
|
31.156
|
NM_001042752
NM_008684
|
Neo1
|
neogenin
|
|
chr5_-_36022193
|
31.062
|
|
Htra3
|
HtrA serine peptidase 3
|
|
chr19_-_57271875
|
30.781
|
NM_001103177
|
Ablim1
|
actin-binding LIM protein 1
|
|
chr7_-_4116300
|
30.768
|
NM_021454
|
Cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chr1_+_137662648
|
30.520
|
NM_013750
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3
|
|
chr6_+_29383152
|
30.122
|
NM_001081185
|
Flnc
|
filamin C, gamma
|
|
chr17_-_35838974
|
29.782
|
NM_001198831
|
Ddr1
|
discoidin domain receptor family, member 1
|
|
chr7_-_133606586
|
29.678
|
NM_007504
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr19_+_5741862
|
29.419
|
|
Ltbp3
|
latent transforming growth factor beta binding protein 3
|
|
chr11_+_87394647
|
29.098
|
|
Sept4
|
septin 4
|
|
chr9_-_8004450
|
28.956
|
|
Yap1
|
yes-associated protein 1
|
|
chr7_-_77511607
|
28.847
|
NM_183261
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr7_-_39012977
|
28.282
|
NM_024413
|
Plekhf1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1
|
|
chr9_-_91260597
|
28.259
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr6_+_124462696
|
27.727
|
|
C1ra
|
complement component 1, r subcomponent A
|
|
chr9_-_91260619
|
27.462
|
NM_009573
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chr11_-_55223449
|
27.056
|
|
Sparc
|
secreted acidic cysteine rich glycoprotein
|
|
chr1_-_170362388
|
26.418
|
NM_008783
NM_183355
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr9_+_78104965
|
25.851
|
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya)
|
|
chr16_-_85080242
|
25.709
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr15_+_98465193
|
25.631
|
NM_007581
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr10_+_21713735
|
25.575
|
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chrX_+_137212566
|
25.509
|
NM_011845
|
Mid2
|
midline 2
|
|
chr12_+_87087488
|
25.081
|
NM_145447
|
Mfsd7c
|
major facilitator superfamily domain containing 7C
|
|
chr2_-_25324650
|
24.751
|
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr6_+_113408477
|
24.703
|
NM_145826
|
Il17re
|
interleukin 17 receptor E
|
|
chr14_-_55196138
|
24.375
|
NM_010590
|
Jub
|
ajuba
|
|
chr7_-_4116102
|
24.070
|
|
Cdc42ep5
|
CDC42 effector protein (Rho GTPase binding) 5
|
|
chrX_-_73119905
|
24.057
|
NM_145629
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr1_+_137732961
|
23.928
|
NM_001130174
NM_001130175
NM_001130177
NM_001130178
NM_001130179
NM_001130180
NM_001130181
NM_011619
NM_001136083
|
Tnnt2
|
troponin T2, cardiac
|
|
chr7_-_51562263
|
23.917
|
NM_009131
|
Clec11a
|
C-type lectin domain family 11, member a
|
|
chr6_+_122463663
|
23.916
|
NM_015776
|
Mfap5
|
microfibrillar associated protein 5
|
|
chr1_+_40323430
|
23.773
|
NM_001123382
|
Il1r1
|
interleukin 1 receptor, type I
|
|
chr9_-_107511523
|
23.687
|
NM_001042779
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B
|
|
chr10_-_22539906
|
23.614
|
NM_011545
|
Tcf21
|
transcription factor 21
|
|
chr3_+_30894668
|
23.417
|
NM_008857
|
Prkci
|
protein kinase C, iota
|
|
chr1_+_40323512
|
23.363
|
|
Il1r1
|
interleukin 1 receptor, type I
|
|
chr8_+_14911653
|
23.342
|
NM_001037736
NM_172751
|
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10
|
|
chr10_-_22539936
|
23.206
|
|
Tcf21
|
transcription factor 21
|
|
chr9_+_72596925
|
23.161
|
|
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4
|
|
chr3_+_90341208
|
23.003
|
|
S100a16
|
S100 calcium binding protein A16
|
|
chr5_-_135422780
|
22.574
|
|
Cldn4
|
claudin 4
|
|
chr9_+_78078492
|
22.389
|
|
Gsta1
|
glutathione S-transferase, alpha 1 (Ya)
|
|
chr7_+_121184667
|
22.193
|
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein
|
|
chr2_+_128689667
|
22.086
|
NM_024237
|
Fbln7
|
fibulin 7
|
|
chr4_-_118216327
|
22.060
|
NM_172383
|
Tmem125
|
transmembrane protein 125
|
|
chr13_+_113254277
|
21.920
|
NM_010560
|
Il6st
|
interleukin 6 signal transducer
|
|
chr19_-_42203380
|
21.855
|
NM_027106
|
Avpi1
|
arginine vasopressin-induced 1
|
|
chr15_-_75839859
|
21.766
|
NM_001168253
|
Fam83h
|
family with sequence similarity 83, member H
|
|
chr6_-_128093543
|
21.741
|
NM_175414
|
Tspan9
|
tetraspanin 9
|
|
chrX_+_137909951
|
21.019
|
NM_001163155
NM_007736
|
Col4a5
|
collagen, type IV, alpha 5
|
|
chr4_+_140857147
|
20.944
|
NM_010139
|
Epha2
|
Eph receptor A2
|
|
chr5_-_135422730
|
20.269
|
|
Cldn4
|
claudin 4
|
|
chr5_-_107718388
|
20.255
|
|
Tgfbr3
|
transforming growth factor, beta receptor III
|
|
chr16_-_38800293
|
20.093
|
NM_178924
|
Upk1b
|
uroplakin 1B
|
|
chr10_+_27794625
|
20.046
|
NM_008983
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K
|
|
chr2_+_160706405
|
19.960
|
NM_022883
|
Lpin3
|
lipin 3
|
|
chr11_-_69211724
|
19.909
|
NM_025915
|
Tmem88
|
transmembrane protein 88
|
|
chr10_+_21713669
|
19.698
|
NM_001161850
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr12_-_99975614
|
19.628
|
NM_001146199
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21
|
|
chr11_+_94797565
|
19.537
|
NM_007742
|
Col1a1
|
collagen, type I, alpha 1
|
|
chr4_+_98213324
|
19.109
|
NM_001005787
NM_007704
|
Inadl
|
InaD-like (Drosophila)
|
|
chr1_+_191552133
|
18.933
|
NM_008976
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14
|
|
chrX_-_73119876
|
18.904
|
|
Pls3
|
plastin 3 (T-isoform)
|
|
chr18_+_23573908
|
18.829
|
NM_010087
NM_207650
|
Dtna
|
dystrobrevin alpha
|
|
chr16_+_38362258
|
18.739
|
NM_001081984
NM_022318
|
Popdc2
|
popeye domain containing 2
|
|
chr7_-_51780003
|
18.733
|
NM_146189
|
Mybpc2
|
myosin binding protein C, fast-type
|
|
chr2_+_119063090
|
18.506
|
NM_016907
|
Spint1
|
serine protease inhibitor, Kunitz type 1
|
|
chr14_-_55613384
|
18.391
|
NM_080728
|
Myh7
|
myosin, heavy polypeptide 7, cardiac muscle, beta
|
|
chr2_+_119063134
|
18.346
|
|
Spint1
|
serine protease inhibitor, Kunitz type 1
|
|
chr11_+_115836022
|
18.164
|
NM_001005608
NM_133663
|
Itgb4
|
integrin beta 4
|
|
chr2_+_20441139
|
17.629
|
NM_001177630
NM_001177631
|
Etl4
|
enhancer trap locus 4
|
|
chr11_-_109472694
|
17.604
|
|
Wipi1
|
WD repeat domain, phosphoinositide interacting 1
|
|
chr8_+_23868194
|
17.466
|
NM_008872
|
Plat
|
plasminogen activator, tissue
|
|
chr8_-_64238814
|
17.436
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr7_-_20334650
|
17.383
|
|
Pvrl2
|
poliovirus receptor-related 2
|
|
chr5_+_136363810
|
17.329
|
|
Hspb1
|
heat shock protein 1
|
|
chr2_-_164683125
|
17.169
|
NM_011125
|
Pltp
|
phospholipid transfer protein
|
|
chr12_+_53604952
|
17.015
|
|
Arhgap5
|
Rho GTPase activating protein 5
|
|
chr9_-_48413181
|
16.988
|
NM_010924
|
Nnmt
|
nicotinamide N-methyltransferase
|
|
chr12_+_8980697
|
16.877
|
NM_001159527
NM_172470
|
Wdr35
|
WD repeat domain 35
|
|
chr6_+_112223751
|
16.877
|
NM_144799
|
Lmcd1
|
LIM and cysteine-rich domains 1
|
|
chr8_+_88364729
|
16.843
|
NM_199446
|
Phkb
|
phosphorylase kinase beta
|
|
chr7_-_29157512
|
16.747
|
NM_001083912
NM_138752
|
Plekhg2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2
|
|
chr15_+_98465328
|
16.479
|
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr10_-_75106702
|
16.404
|
NM_001162913
NM_027890
|
Susd2
|
sushi domain containing 2
|
|
chr17_-_46162111
|
16.290
|
NM_001110266
NM_001110267
NM_001110268
|
Vegfa
|
vascular endothelial growth factor A
|
|
chr6_+_124462638
|
15.957
|
NM_023143
|
C1ra
|
complement component 1, r subcomponent A
|
|
chr19_-_40525876
|
15.862
|
NM_001034962
NM_001034963
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr18_+_23573939
|
15.842
|
|
Dtna
|
dystrobrevin alpha
|
|
chr3_-_57379831
|
15.778
|
NM_001168281
|
Wwtr1
|
WW domain containing transcription regulator 1
|
|
chr10_+_27794664
|
15.678
|
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K
|
|
chr2_+_119063436
|
15.508
|
|
Spint1
|
serine protease inhibitor, Kunitz type 1
|
|
chr18_-_12464213
|
15.484
|
NM_001190371
|
Ankrd29
|
ankyrin repeat domain 29
|
|
chr12_+_30687597
|
15.453
|
|
Pxdn
|
peroxidasin homolog (Drosophila)
|
|
chr18_-_12464173
|
15.357
|
|
Ankrd29
|
ankyrin repeat domain 29
|
|
chr5_+_48374328
|
14.984
|
NM_178804
|
Slit2
|
slit homolog 2 (Drosophila)
|
|
chr5_+_104888468
|
14.686
|
NM_008861
|
Pkd2
|
polycystic kidney disease 2
|
|
chr10_+_128370814
|
14.615
|
NM_008398
|
Itga7
|
integrin alpha 7
|
|
chr5_-_135422795
|
14.422
|
NM_009903
|
Cldn4
|
claudin 4
|
|
chr9_+_46036788
|
14.376
|
|
Apoa1
|
apolipoprotein A-I
|
|
chr2_+_119063194
|
14.342
|
|
|
|
|
chr4_+_133871031
|
14.306
|
NM_001039048
|
Trim63
|
tripartite motif-containing 63
|
|
chr4_+_57858096
|
14.220
|
NM_001035532
NM_009649
|
Akap2
|
A kinase (PRKA) anchor protein 2
|
|
chr7_-_77511242
|
14.220
|
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2
|
|
chr2_-_25325196
|
14.209
|
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr8_-_67171805
|
14.044
|
NM_013494
|
Cpe
|
carboxypeptidase E
|
|
chr5_+_136363914
|
13.828
|
|
Hspb1
|
heat shock protein 1
|
|
chr18_+_23573977
|
13.736
|
|
Dtna
|
dystrobrevin alpha
|
|
chr1_-_136700389
|
13.452
|
|
Ppp1r12b
|
protein phosphatase 1, regulatory (inhibitor) subunit 12B
|
|
chr16_-_31314534
|
13.272
|
NM_007470
|
Apod
|
apolipoprotein D
|
|
chr9_-_48413097
|
13.146
|
|
Nnmt
|
nicotinamide N-methyltransferase
|
|
chr16_+_37777075
|
13.102
|
NM_008047
|
Fstl1
|
follistatin-like 1
|
|
chr8_-_75877407
|
12.907
|
|
Large
|
like-glycosyltransferase
|
|
chr6_-_37391993
|
12.905
|
NM_178661
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2
|
|
chr8_-_13494457
|
12.723
|
NM_019521
|
Gas6
|
growth arrest specific 6
|
|
chr5_-_36022114
|
12.690
|
|
Htra3
|
HtrA serine peptidase 3
|
|
chr1_-_89291303
|
12.658
|
NM_001110227
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13
|
|
chr2_-_6134008
|
12.584
|
NM_024208
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3
|
|
chr9_-_79566271
|
12.379
|
NM_007730
|
Col12a1
|
collagen, type XII, alpha 1
|
|
chr10_+_21705909
|
12.333
|
NM_001161848
|
Sgk1
|
serum/glucocorticoid regulated kinase 1
|
|
chr11_-_100831819
|
12.214
|
|
Ptrf
|
polymerase I and transcript release factor
|
|
chr19_+_29999624
|
12.170
|
|
Il33
|
interleukin 33
|
|
chr7_-_20284370
|
12.162
|
|
Apoe
|
apolipoprotein E
|
|
chr13_+_81022644
|
12.108
|
NM_001042591
|
Arrdc3
|
arrestin domain containing 3
|
|
chr2_-_25325248
|
12.095
|
NM_008963
|
Ptgds
|
prostaglandin D2 synthase (brain)
|
|
chr14_+_70009282
|
12.027
|
NM_033325
|
Loxl2
|
lysyl oxidase-like 2
|
|
chr14_+_120705667
|
11.804
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr3_+_79690856
|
11.796
|
|
Fam198b
|
family with sequence similarity 198, member B
|
|
chr7_+_97535516
|
11.788
|
|
Sytl2
|
synaptotagmin-like 2
|
|
chr9_+_46037941
|
11.785
|
|
|
|
|
chr2_-_25947915
|
11.585
|
NM_001037098
|
Nacc2
|
nucleus accumbens associated 2, BEN and BTB (POZ) domain containing
|