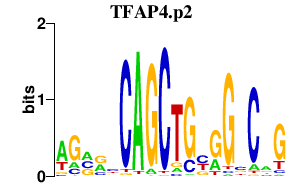
|
chr18_-_64820573
|
12.044
|
NM_001001488
|
Atp8b1
|
ATPase, class I, type 8B, member 1
|
|
chr7_-_30017388
|
11.676
|
NM_001033525
|
Kcnk6
|
potassium inwardly-rectifying channel, subfamily K, member 6
|
|
chr8_-_107995282
|
11.294
|
NM_026481
|
Tppp3
|
tubulin polymerization-promoting protein family member 3
|
|
chr8_-_107995252
|
10.779
|
|
Tppp3
|
tubulin polymerization-promoting protein family member 3
|
|
chr14_-_22806999
|
9.844
|
|
Zfp503
|
zinc finger protein 503
|
|
chr9_-_58160769
|
9.774
|
|
Loxl1
|
lysyl oxidase-like 1
|
|
chr10_-_76172218
|
9.529
|
|
Col6a1
|
collagen, type VI, alpha 1
|
|
chr4_+_104829963
|
9.494
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr10_-_76576422
|
9.240
|
NM_001109991
|
Col18a1
|
collagen, type XVIII, alpha 1
|
|
chr4_+_104830022
|
9.065
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr12_+_76409299
|
9.038
|
NM_023275
|
Rhoj
|
ras homolog gene family, member J
|
|
chr4_+_104829951
|
9.014
|
NM_080555
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr3_+_90345082
|
8.954
|
NM_026416
|
S100a16
|
S100 calcium binding protein A16
|
|
chr11_-_32122236
|
8.661
|
|
Rhbdf1
|
rhomboid family 1 (Drosophila)
|
|
chr8_-_11312729
|
8.571
|
NM_009931
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr1_-_193141946
|
8.483
|
NM_025424
|
Nenf
|
neuron derived neurotrophic factor
|
|
chr7_+_80885223
|
8.090
|
|
Fam174b
|
family with sequence similarity 174, member B
|
|
chr4_+_104829867
|
8.016
|
|
Ppap2b
|
phosphatidic acid phosphatase type 2B
|
|
chr11_+_69778998
|
7.822
|
|
Cldn7
|
claudin 7
|
|
chr11_+_69778907
|
7.812
|
NM_016887
|
Cldn7
|
claudin 7
|
|
chr7_+_138079669
|
7.706
|
NM_019564
|
Htra1
|
HtrA serine peptidase 1
|
|
chr4_-_117963950
|
7.676
|
NM_011213
|
Ptprf
|
protein tyrosine phosphatase, receptor type, F
|
|
chr8_-_67171481
|
7.646
|
|
Cpe
|
carboxypeptidase E
|
|
chr19_-_24936327
|
7.543
|
NM_175013
|
Pgm5
|
phosphoglucomutase 5
|
|
chr8_-_11312686
|
7.404
|
|
Col4a1
|
collagen, type IV, alpha 1
|
|
chr10_-_94877824
|
7.345
|
NM_001168657
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr1_+_44608355
|
7.199
|
NM_028450
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1
|
|
chr11_+_101192080
|
7.189
|
|
Aoc3
|
amine oxidase, copper containing 3
|
|
chr10_-_94878117
|
7.150
|
NM_001168656
|
Socs2
|
suppressor of cytokine signaling 2
|
|
chr2_-_26459777
|
6.900
|
|
Agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta)
|
|
chr3_-_151929210
|
6.793
|
NM_199465
|
Nexn
|
nexilin
|
|
chr8_-_67171805
|
6.752
|
NM_013494
|
Cpe
|
carboxypeptidase E
|
|
chr2_+_35139479
|
6.688
|
|
Gsn
|
gelsolin
|
|
chr11_-_3427914
|
6.678
|
|
Smtn
|
smoothelin
|
|
chr6_+_4465627
|
6.565
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr10_-_116719183
|
6.565
|
NM_017372
|
Lyz2
|
lysozyme 2
|
|
chr2_-_26459710
|
6.534
|
NM_026212
|
Agpat2
|
1-acylglycerol-3-phosphate O-acyltransferase 2 (lysophosphatidic acid acyltransferase, beta)
|
|
chr6_+_56782073
|
6.519
|
|
Fkbp9
|
FK506 binding protein 9
|
|
chr1_-_195099209
|
6.423
|
|
G0s2
|
G0/G1 switch gene 2
|
|
chr10_-_116719364
|
6.409
|
|
Lyz2
|
lysozyme 2
|
|
chr7_+_35905996
|
6.396
|
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha
|
|
chr14_+_70009282
|
6.374
|
NM_033325
|
Loxl2
|
lysyl oxidase-like 2
|
|
chr11_+_61298923
|
6.356
|
NM_029568
|
Mfap4
|
microfibrillar-associated protein 4
|
|
chr14_-_22806631
|
6.352
|
|
Zfp503
|
zinc finger protein 503
|
|
chr6_+_43215604
|
6.170
|
NM_133674
|
Arhgef5
|
Rho guanine nucleotide exchange factor (GEF) 5
|
|
chr18_-_53577625
|
5.989
|
NM_008908
|
Ppic
|
peptidylprolyl isomerase C
|
|
chrX_-_109812258
|
5.926
|
NM_181579
|
Pof1b
|
premature ovarian failure 1B
|
|
chr2_+_155207542
|
5.869
|
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2
|
|
chr19_-_40588193
|
5.869
|
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr9_-_58160063
|
5.766
|
|
Loxl1
|
lysyl oxidase-like 1
|
|
chr4_-_43536296
|
5.757
|
|
Tpm2
|
tropomyosin 2, beta
|
|
chr4_+_140976693
|
5.745
|
NM_013868
|
Hspb7
|
heat shock protein family, member 7 (cardiovascular)
|
|
chr2_-_165981138
|
5.732
|
NM_028072
|
Sulf2
|
sulfatase 2
|
|
chr2_+_32731648
|
5.700
|
NM_146119
|
Fam129b
|
family with sequence similarity 129, member B
|
|
chr3_+_104591946
|
5.689
|
NM_007484
|
Rhoc
|
ras homolog gene family, member C
|
|
chr13_+_56710896
|
5.656
|
NM_009369
|
Tgfbi
|
transforming growth factor, beta induced
|
|
chr9_+_50561315
|
5.596
|
|
Cryab
|
crystallin, alpha B
|
|
chr16_+_14705932
|
5.558
|
NM_011415
|
Snai2
|
snail homolog 2 (Drosophila)
|
|
chr19_-_40588235
|
5.505
|
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr9_+_53192102
|
5.497
|
NM_212445
|
Kdelc2
|
KDEL (Lys-Asp-Glu-Leu) containing 2
|
|
chr2_-_165981085
|
5.465
|
|
Sulf2
|
sulfatase 2
|
|
chr10_-_116729893
|
5.440
|
NM_013590
|
Lyz1
|
lysozyme 1
|
|
chr11_-_7113899
|
5.430
|
NM_008343
|
Igfbp3
|
insulin-like growth factor binding protein 3
|
|
chr2_+_145611851
|
5.408
|
NM_028724
|
Rin2
|
Ras and Rab interactor 2
|
|
chr19_-_40588219
|
5.391
|
NM_178362
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr15_-_58046380
|
5.374
|
|
Fbxo32
|
F-box protein 32
|
|
chr13_+_23776066
|
5.337
|
NM_023422
|
Hist1h2bc
|
histone cluster 1, H2bc
|
|
chr16_+_45094196
|
5.309
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr3_+_104591955
|
5.276
|
|
Rhoc
|
ras homolog gene family, member C
|
|
chr12_-_103057112
|
5.267
|
|
Fbln5
|
fibulin 5
|
|
chr8_-_64069935
|
5.236
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr11_-_68200284
|
5.212
|
NM_008744
|
Ntn1
|
netrin 1
|
|
chr14_+_34736772
|
5.212
|
NM_145741
|
Gdf10
|
growth differentiation factor 10
|
|
chr10_+_87322774
|
5.192
|
|
Igf1
|
insulin-like growth factor 1
|
|
chr18_+_20716598
|
5.160
|
NM_007883
|
Dsg2
|
desmoglein 2
|
|
chr13_-_23802696
|
5.153
|
|
Hfe
|
hemochromatosis
|
|
chr19_-_40588297
|
5.128
|
NM_009166
|
Sorbs1
|
sorbin and SH3 domain containing 1
|
|
chr8_-_42459932
|
5.100
|
NM_019734
|
Asah1
|
N-acylsphingosine amidohydrolase 1
|
|
chr18_+_60963495
|
5.083
|
NM_001042605
NM_010545
|
Cd74
|
CD74 antigen (invariant polypeptide of major histocompatibility complex, class II antigen-associated)
|
|
chr17_-_25218058
|
5.031
|
NM_001001183
|
Tmem204
|
transmembrane protein 204
|
|
chr7_-_26601526
|
4.998
|
NM_028775
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1
|
|
chr9_+_50560862
|
4.924
|
NM_009964
|
Cryab
|
crystallin, alpha B
|
|
chr4_-_11313886
|
4.857
|
NM_194055
|
Esrp1
|
epithelial splicing regulatory protein 1
|
|
chr4_-_42393043
|
4.848
|
NM_011335
NM_001193668
NM_001193666
NM_023052
|
Ccl21b
Gm10591
Gm13304
Ccl21a
Ccl21c
|
chemokine (C-C motif) ligand 21B (leucine)
predicted gene 10591
predicted gene 13304
chemokine (C-C motif) ligand 21A (serine)
chemokine (C-C motif) ligand 21C (leucine)
|
|
chr14_+_34736806
|
4.764
|
|
Gdf10
|
growth differentiation factor 10
|
|
chr13_+_56711111
|
4.756
|
|
Tgfbi
|
transforming growth factor, beta induced
|
|
chr10_+_69707847
|
4.683
|
|
Slc16a9
|
solute carrier family 16 (monocarboxylic acid transporters), member 9
|
|
chr16_+_45094214
|
4.667
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr8_-_67171692
|
4.660
|
|
Cpe
|
carboxypeptidase E
|
|
chr7_-_139346304
|
4.654
|
|
Cpxm2
|
carboxypeptidase X 2 (M14 family)
|
|
chr15_+_34167780
|
4.642
|
NM_033521
|
Laptm4b
|
lysosomal-associated protein transmembrane 4B
|
|
chr9_+_50561121
|
4.628
|
|
Cryab
|
crystallin, alpha B
|
|
chr4_-_136448748
|
4.597
|
NM_007574
|
C1qc
|
complement component 1, q subcomponent, C chain
|
|
chr5_-_50450152
|
4.577
|
NM_133911
|
Gpr125
|
G protein-coupled receptor 125
|
|
chr8_-_75876420
|
4.525
|
NM_010687
|
Large
|
like-glycosyltransferase
|
|
chr2_+_102390226
|
4.481
|
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr3_+_27837540
|
4.478
|
NM_001164056
|
Pld1
|
phospholipase D1
|
|
chr9_+_43942734
|
4.431
|
NM_023061
|
Mcam
|
melanoma cell adhesion molecule
|
|
chr2_+_33071451
|
4.411
|
NM_011923
|
Angptl2
|
angiopoietin-like 2
|
|
chr2_+_102390177
|
4.332
|
NM_173749
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1
|
|
chr7_-_149763600
|
4.322
|
|
H19
|
H19 fetal liver mRNA
|
|
chr10_+_79353597
|
4.306
|
NM_013459
|
Cfd
|
complement factor D (adipsin)
|
|
chr1_-_155179844
|
4.296
|
NM_010683
|
Lamc1
|
laminin, gamma 1
|
|
chr2_-_181406313
|
4.294
|
NM_009236
|
Sox18
|
SRY-box containing gene 18
|
|
chr6_-_125444695
|
4.259
|
|
Cd9
|
CD9 antigen
|
|
chr9_+_65478364
|
4.254
|
NM_028030
|
Rbpms2
|
RNA binding protein with multiple splicing 2
|
|
chr8_+_128519001
|
4.229
|
NM_008430
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr4_-_43536259
|
4.221
|
NM_009416
|
Tpm2
|
tropomyosin 2, beta
|
|
chr1_+_91351435
|
4.215
|
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr7_+_3290587
|
4.213
|
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr1_-_195099285
|
4.204
|
NM_008059
|
G0s2
|
G0/G1 switch gene 2
|
|
chr7_-_26588561
|
4.170
|
|
Cyp2s1
|
cytochrome P450, family 2, subfamily s, polypeptide 1
|
|
chr19_+_44368557
|
4.161
|
|
Scd2
|
stearoyl-Coenzyme A desaturase 2
|
|
chr7_-_135039020
|
4.160
|
NM_178600
|
Vkorc1
|
vitamin K epoxide reductase complex, subunit 1
|
|
chr15_-_76447278
|
4.135
|
NM_028064
|
Slc39a4
|
solute carrier family 39 (zinc transporter), member 4
|
|
chr8_-_94325138
|
4.126
|
NM_008393
|
Irx3
|
Iroquois related homeobox 3 (Drosophila)
|
|
chr7_-_20282950
|
4.082
|
|
Apoe
|
apolipoprotein E
|
|
chr17_-_57367499
|
4.067
|
|
C3
|
complement component 3
|
|
chr6_-_39068241
|
4.056
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr9_+_50561381
|
4.019
|
|
Cryab
|
crystallin, alpha B
|
|
chr7_-_148086285
|
3.987
|
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb
|
|
chr7_+_29618812
|
3.982
|
NM_010706
|
Lgals4
Lgals6
|
lectin, galactose binding, soluble 4
lectin, galactose binding, soluble 6
|
|
chr9_+_23027519
|
3.922
|
NM_028472
|
Bmper
|
BMP-binding endothelial regulator
|
|
chr9_+_50561092
|
3.914
|
|
Cryab
|
crystallin, alpha B
|
|
chr19_+_44368578
|
3.890
|
|
Scd2
|
stearoyl-Coenzyme A desaturase 2
|
|
chr2_+_35122282
|
3.882
|
|
Gsn
|
gelsolin
|
|
chr11_+_76015590
|
3.877
|
|
Fam57a
|
family with sequence similarity 57, member A
|
|
chr12_-_103056971
|
3.858
|
|
Fbln5
|
fibulin 5
|
|
chr16_+_15887375
|
3.846
|
NM_007679
|
Cebpd
|
CCAAT/enhancer binding protein (C/EBP), delta
|
|
chr11_-_118216591
|
3.834
|
|
Timp2
|
tissue inhibitor of metalloproteinase 2
|
|
chr6_+_88674821
|
3.821
|
|
Mgll
|
monoglyceride lipase
|
|
chr6_-_39068340
|
3.821
|
NM_172893
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr19_+_44368576
|
3.729
|
|
Scd2
|
stearoyl-Coenzyme A desaturase 2
|
|
chr2_-_165980462
|
3.726
|
|
Sulf2
|
sulfatase 2
|
|
chr2_-_76511070
|
3.682
|
NM_010222
|
Fkbp7
|
FK506 binding protein 7
|
|
chr17_-_57367542
|
3.678
|
NM_009778
|
C3
|
complement component 3
|
|
chr19_+_38470458
|
3.666
|
NM_175507
|
Tmem20
|
transmembrane protein 20
|
|
chr7_+_148661329
|
3.665
|
|
Tspan4
|
tetraspanin 4
|
|
chr8_+_128519295
|
3.657
|
|
Kcnk1
|
potassium channel, subfamily K, member 1
|
|
chr5_+_35384200
|
3.653
|
NM_019447
|
Hgfac
|
hepatocyte growth factor activator
|
|
chr15_+_101242832
|
3.632
|
NM_033073
|
Krt7
|
keratin 7
|
|
chr7_+_3290516
|
3.613
|
NM_016969
NM_001093765
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr2_-_13977502
|
3.583
|
|
Ptpla
|
protein tyrosine phosphatase-like (proline instead of catalytic arginine), member a
|
|
chr17_+_15080188
|
3.574
|
NM_001083880
NM_001083881
NM_001083882
NM_026451
|
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene
|
|
chr16_+_45094383
|
3.567
|
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr18_+_58038278
|
3.563
|
NM_009194
|
Slc12a2
|
solute carrier family 12, member 2
|
|
chr10_-_27336648
|
3.559
|
NM_008481
|
Lama2
|
laminin, alpha 2
|
|
chr6_-_39067918
|
3.555
|
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12
|
|
chr4_-_134828503
|
3.555
|
NM_013885
|
Clic4
|
chloride intracellular channel 4 (mitochondrial)
|
|
chr5_+_17080553
|
3.496
|
NM_013657
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr14_-_31981750
|
3.482
|
|
Stab1
|
stabilin 1
|
|
chr10_-_60610398
|
3.463
|
NM_009163
|
Sgpl1
|
sphingosine phosphate lyase 1
|
|
chr4_-_57155985
|
3.462
|
|
Epb4.1l4b
|
erythrocyte protein band 4.1-like 4b
|
|
chr1_+_91351373
|
3.457
|
NM_001037136
NM_178119
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chrX_+_165233030
|
3.456
|
NM_009707
|
Arhgap6
|
Rho GTPase activating protein 6
|
|
chr3_-_88351996
|
3.449
|
NM_016899
|
Rab25
|
RAB25, member RAS oncogene family
|
|
chr14_+_33669941
|
3.428
|
NM_178791
|
E130203B14Rik
|
RIKEN cDNA E130203B14 gene
|
|
chr8_+_28370796
|
3.414
|
NM_007918
|
Eif4ebp1
|
eukaryotic translation initiation factor 4E binding protein 1
|
|
chr2_+_16277963
|
3.406
|
|
Plxdc2
|
plexin domain containing 2
|
|
chrX_-_147241307
|
3.400
|
|
Maged2
|
melanoma antigen, family D, 2
|
|
chr15_+_34236408
|
3.389
|
NM_016762
|
Matn2
|
matrilin 2
|
|
chr8_+_71404491
|
3.337
|
|
Lpl
|
lipoprotein lipase
|
|
chr1_-_51972718
|
3.332
|
|
Myo1b
|
myosin IB
|
|
chr16_+_93683426
|
3.300
|
NM_173047
|
Cbr3
|
carbonyl reductase 3
|
|
chr15_-_100467247
|
3.294
|
NM_174992
NM_001033872
|
Smagp
|
small cell adhesion glycoprotein
|
|
chr14_+_33670038
|
3.282
|
|
E130203B14Rik
|
RIKEN cDNA E130203B14 gene
|
|
chr7_-_28118628
|
3.279
|
NM_001113549
|
Ltbp4
|
latent transforming growth factor beta binding protein 4
|
|
chr4_-_136438225
|
3.276
|
|
C1qb
|
complement component 1, q subcomponent, beta polypeptide
|
|
chr13_-_3803547
|
3.275
|
NM_027416
|
Calml3
|
calmodulin-like 3
|
|
chr10_+_76721977
|
3.269
|
NM_030262
|
Pofut2
|
protein O-fucosyltransferase 2
|
|
chr18_+_12492494
|
3.268
|
NM_010680
|
Lama3
|
laminin, alpha 3
|
|
chr7_+_96552871
|
3.255
|
NM_008055
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr9_-_63868311
|
3.225
|
|
Smad6
|
MAD homolog 6 (Drosophila)
|
|
chr17_-_23877115
|
3.208
|
NM_023824
|
Paqr4
|
progestin and adipoQ receptor family member IV
|
|
chr7_-_118923483
|
3.201
|
NM_173739
|
Galntl4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4
|
|
chrX_-_137077133
|
3.179
|
|
Tsc22d3
|
TSC22 domain family, member 3
|
|
chr10_+_23943322
|
3.179
|
NM_021509
|
Moxd1
|
monooxygenase, DBH-like 1
|
|
chr7_-_133606617
|
3.171
|
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr2_-_104334599
|
3.166
|
|
|
|
|
chrX_+_147778717
|
3.134
|
NM_198105
|
Fam120c
|
family with sequence similarity 120, member C
|
|
chr1_-_52557130
|
3.123
|
NM_008667
|
Nab1
|
Ngfi-A binding protein 1
|
|
chr15_+_38908403
|
3.078
|
NM_026778
|
Cthrc1
|
collagen triple helix repeat containing 1
|
|
chr15_+_34167908
|
3.074
|
|
Laptm4b
|
lysosomal-associated protein transmembrane 4B
|
|
chr14_+_70008874
|
3.051
|
|
Loxl2
|
lysyl oxidase-like 2
|
|
chr17_+_29455821
|
3.044
|
NM_023734
|
Pi16
|
peptidase inhibitor 16
|
|
chr7_-_133606586
|
3.039
|
NM_007504
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr7_+_148661138
|
3.034
|
NM_053082
|
Tspan4
|
tetraspanin 4
|
|
chr11_-_3831515
|
3.022
|
|
Tcn2
|
transcobalamin 2
|
|
chr2_-_80421054
|
3.019
|
NM_016965
|
Nckap1
|
NCK-associated protein 1
|
|
chr9_-_58160966
|
3.018
|
NM_010729
|
Loxl1
|
lysyl oxidase-like 1
|
|
chr17_-_33918179
|
3.017
|
|
Angptl4
|
angiopoietin-like 4
|
|
chr17_-_28697643
|
3.005
|
NM_025469
|
Clps
|
colipase, pancreatic
|
|
chr19_-_36811090
|
3.001
|
NM_016854
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C
|
|
chrX_-_151650799
|
2.971
|
|
Sat1
|
spermidine/spermine N1-acetyl transferase 1
|
|
chr1_+_72871060
|
2.966
|
NM_008342
|
Igfbp2
|
insulin-like growth factor binding protein 2
|
|
chr7_+_96552955
|
2.959
|
|
Fzd4
|
frizzled homolog 4 (Drosophila)
|
|
chr10_+_115738429
|
2.952
|
NM_029928
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B
|
|
chr9_-_108166059
|
2.945
|
NM_010017
|
Dag1
|
dystroglycan 1
|
|
chr2_-_6134008
|
2.936
|
NM_024208
|
Echdc3
|
enoyl Coenzyme A hydratase domain containing 3
|
|
chr10_+_12810593
|
2.935
|
NM_009538
|
Plagl1
|
pleiomorphic adenoma gene-like 1
|