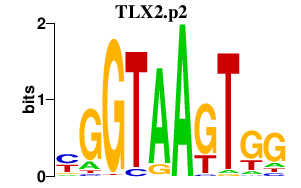
|
chr1_+_45401303
|
20.578
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_+_45401659
|
18.753
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr1_-_45559866
|
12.765
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr1_-_45559904
|
12.359
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr7_+_134355216
|
12.239
|
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr7_+_134355120
|
12.132
|
NM_016754
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle
|
|
chr16_-_56886260
|
11.869
|
NM_019631
|
Tmem45a
|
transmembrane protein 45a
|
|
chr1_-_45559700
|
11.389
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr10_+_79451332
|
11.341
|
NM_007725
|
Cnn2
|
calponin 2
|
|
chr10_+_97028456
|
11.041
|
|
Lum
|
lumican
|
|
chr10_+_79451369
|
10.508
|
|
Cnn2
|
calponin 2
|
|
chr11_-_78798332
|
10.477
|
NM_001159301
NM_010708
|
Lgals9
|
lectin, galactose binding, soluble 9
|
|
chr3_-_92819586
|
10.322
|
NM_028798
|
Crct1
|
cysteine-rich C-terminal 1
|
|
chr1_-_92740348
|
10.264
|
|
Col6a3
|
collagen, type VI, alpha 3
|
|
chr9_-_45743950
|
10.151
|
|
Tagln
|
transgelin
|
|
chr3_+_54165028
|
10.012
|
NM_001198765
NM_001198766
NM_015784
|
Postn
|
periostin, osteoblast specific factor
|
|
chr3_+_90472990
|
9.688
|
NM_013650
|
S100a8
|
S100 calcium binding protein A8 (calgranulin A)
|
|
chr9_-_66880619
|
9.335
|
|
Tpm1
|
tropomyosin 1, alpha
|
|
chr8_+_95351212
|
9.185
|
NM_008610
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr9_-_45744036
|
9.028
|
NM_011526
|
Tagln
|
transgelin
|
|
chr7_-_149762919
|
9.004
|
|
H19
|
H19 fetal liver mRNA
|
|
chr1_-_45560001
|
8.889
|
NM_007737
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr8_+_95376459
|
8.740
|
|
Mmp2
|
matrix metallopeptidase 2
|
|
chr10_+_97028409
|
8.118
|
|
Lum
|
lumican
|
|
chr3_+_90407688
|
8.013
|
NM_011311
|
S100a4
|
S100 calcium binding protein A4
|
|
chr1_+_45368443
|
8.011
|
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr3_+_92483168
|
7.861
|
NM_028622
|
Lce1c
|
late cornified envelope 1C
|
|
chr2_-_84614976
|
7.731
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr3_-_92474213
|
7.590
|
NM_028625
|
Lce1a2
|
late cornified envelope 1A2
|
|
chr6_-_124586102
|
7.582
|
NM_173864
|
Gm5077
|
predicted gene 5077
|
|
chr15_+_54577256
|
7.549
|
NM_010930
|
Nov
|
nephroblastoma overexpressed gene
|
|
chr2_-_84614960
|
7.422
|
|
Serping1
|
serine (or cysteine) peptidase inhibitor, clade G, member 1
|
|
chr12_+_25393083
|
7.373
|
NM_009104
|
Rrm2
|
ribonucleotide reductase M2
|
|
chr8_-_26212588
|
7.321
|
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2
|
|
chr9_-_21935019
|
7.109
|
NM_007388
|
Acp5
|
acid phosphatase 5, tartrate resistant
|
|
chr7_-_133606586
|
7.100
|
NM_007504
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr1_-_45460310
|
7.071
|
|
Col5a2
|
collagen, type V, alpha 2
|
|
chr19_-_34329804
|
7.036
|
NM_007392
|
Acta2
|
actin, alpha 2, smooth muscle, aorta
|
|
chr7_-_125629996
|
6.956
|
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr7_-_133606617
|
6.840
|
|
Atp2a1
|
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
|
|
chr3_+_93123668
|
6.839
|
NM_133698
|
Hrnr
|
hornerin
|
|
chrX_+_156146532
|
6.542
|
NM_021389
|
Sh3kbp1
|
SH3-domain kinase binding protein 1
|
|
chr1_-_45982242
|
6.315
|
NM_016917
|
Slc40a1
|
solute carrier family 40 (iron-regulated transporter), member 1
|
|
chr6_+_122463595
|
6.299
|
|
Mfap5
|
microfibrillar associated protein 5
|
|
chr6_+_4471231
|
6.281
|
|
Col1a2
|
collagen, type I, alpha 2
|
|
chr8_+_11443165
|
6.267
|
|
Col4a2
|
collagen, type IV, alpha 2
|
|
chr8_-_64238843
|
6.168
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr13_-_74946095
|
6.164
|
|
Cast
|
calpastatin
|
|
chr1_+_45368382
|
6.141
|
NM_009930
|
Col3a1
|
collagen, type III, alpha 1
|
|
chr16_-_45844409
|
6.120
|
NM_153412
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr6_+_135015679
|
6.105
|
NM_181444
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A
|
|
chr8_-_64238814
|
6.102
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr4_-_63707849
|
6.026
|
|
Tnc
|
tenascin C
|
|
chr19_-_5924796
|
5.983
|
|
Cdc42ep2
|
CDC42 effector protein (Rho GTPase binding) 2
|
|
chr7_-_53176582
|
5.895
|
NM_010129
|
Emp3
|
epithelial membrane protein 3
|
|
chr3_-_92452189
|
5.748
|
NM_025984
|
Lce1a1
|
late cornified envelope 1A1
|
|
chr2_-_164182009
|
5.717
|
|
Slpi
|
secretory leukocyte peptidase inhibitor
|
|
chr11_-_83391984
|
5.695
|
|
Ccl9
|
chemokine (C-C motif) ligand 9
|
|
chr6_-_136890189
|
5.684
|
NM_007486
|
Arhgdib
|
Rho, GDP dissociation inhibitor (GDI) beta
|
|
chr16_-_10314010
|
5.674
|
NM_007929
|
Emp2
|
epithelial membrane protein 2
|
|
chr16_-_10313963
|
5.651
|
|
Emp2
|
epithelial membrane protein 2
|
|
chr3_-_92582820
|
5.592
|
NM_029667
|
Lce1i
|
late cornified envelope 1I
|
|
chr1_-_71699671
|
5.402
|
NM_010233
|
Fn1
|
fibronectin 1
|
|
chr6_+_122463663
|
5.322
|
NM_015776
|
Mfap5
|
microfibrillar associated protein 5
|
|
chr3_+_92120626
|
5.307
|
NM_011469
|
Sprr2b
|
small proline-rich protein 2B
|
|
chr6_+_117118552
|
5.259
|
NM_001012477
NM_013655
NM_021704
|
Cxcl12
|
chemokine (C-X-C motif) ligand 12
|
|
chr9_+_110921791
|
5.231
|
NM_008522
|
Ltf
|
lactotransferrin
|
|
chr7_-_56103405
|
5.178
|
NM_001198841
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr18_-_52688607
|
5.170
|
|
Lox
|
lysyl oxidase
|
|
chr10_-_90545046
|
5.160
|
|
Apaf1
|
apoptotic peptidase activating factor 1
|
|
chr3_+_90318894
|
5.059
|
|
S100a13
|
S100 calcium binding protein A13
|
|
chr4_+_118104028
|
5.038
|
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr11_-_54798601
|
5.007
|
|
|
|
|
chr10_-_117261663
|
4.954
|
|
Rap1b
|
RAS related protein 1b
|
|
chr12_+_114378688
|
4.925
|
|
Crip2
|
cysteine rich protein 2
|
|
chr3_-_92524255
|
4.906
|
NM_026394
|
Lce1f
|
late cornified envelope 1F
|
|
chr3_+_90318850
|
4.904
|
|
S100a13
|
S100 calcium binding protein A13
|
|
chr7_-_56103389
|
4.887
|
|
Csrp3
|
cysteine and glycine-rich protein 3
|
|
chr17_-_35973192
|
4.875
|
|
Tubb5
|
tubulin, beta 5
|
|
chr11_+_82930294
|
4.805
|
NM_011407
|
Slfn1
|
schlafen 1
|
|
chr11_-_75841236
|
4.752
|
NM_029658
|
Fam101b
|
family with sequence similarity 101, member B
|
|
chr2_+_164595447
|
4.728
|
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr14_+_34899165
|
4.725
|
NM_013473
|
Anxa8
|
annexin A8
|
|
chr16_+_24721940
|
4.678
|
NM_001145954
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr7_+_135390325
|
4.658
|
NM_009365
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr5_+_17080553
|
4.641
|
NM_013657
|
Sema3c
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C
|
|
chr10_+_79321979
|
4.632
|
|
Ptbp1
|
polypyrimidine tract binding protein 1
|
|
chr9_+_7764067
|
4.620
|
NM_133739
|
Tmem123
|
transmembrane protein 123
|
|
chr11_+_45869506
|
4.600
|
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chr19_-_46119403
|
4.556
|
|
Ldb1
|
LIM domain binding 1
|
|
chr11_+_61298923
|
4.504
|
NM_029568
|
Mfap4
|
microfibrillar-associated protein 4
|
|
chr2_+_164595415
|
4.499
|
NM_026785
|
Ube2c
|
ubiquitin-conjugating enzyme E2C
|
|
chr12_+_8778260
|
4.476
|
|
Sdc1
|
syndecan 1
|
|
chr17_+_34400152
|
4.468
|
NM_207105
|
H2-Ab1
|
histocompatibility 2, class II antigen A, beta 1
|
|
chr5_-_138312923
|
4.411
|
NM_001024932
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2
|
|
chr11_-_106576380
|
4.380
|
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1
|
|
chr11_+_104469313
|
4.377
|
NM_016780
|
Itgb3
|
integrin beta 3
|
|
chr7_-_51787440
|
4.373
|
NM_019866
|
Spib
|
Spi-B transcription factor (Spi-1/PU.1 related)
|
|
chr17_-_12888210
|
4.364
|
|
Igf2r
|
insulin-like growth factor 2 receptor
|
|
chr9_+_62705890
|
4.352
|
NM_001102468
NM_138304
|
Calml4
|
calmodulin-like 4
|
|
chr1_-_132359220
|
4.345
|
|
Cd55
|
CD55 antigen
|
|
chr7_-_133512962
|
4.340
|
NM_010689
|
Lat
|
linker for activation of T cells
|
|
chr9_-_70269326
|
4.333
|
NM_007630
|
Ccnb2
|
cyclin B2
|
|
chr17_-_57367499
|
4.294
|
|
C3
|
complement component 3
|
|
chr3_+_87752614
|
4.277
|
NM_007759
|
Crabp2
|
cellular retinoic acid binding protein II
|
|
chr7_+_135390456
|
4.277
|
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1
|
|
chr2_-_72817058
|
4.273
|
|
Sp3
|
trans-acting transcription factor 3
|
|
chr8_-_64238879
|
4.271
|
|
Palld
|
palladin, cytoskeletal associated protein
|
|
chr6_-_99216494
|
4.258
|
NM_001197321
|
Foxp1
|
forkhead box P1
|
|
chr11_+_61298978
|
4.256
|
|
Mfap4
|
microfibrillar-associated protein 4
|
|
chr10_+_97028134
|
4.246
|
NM_008524
|
Lum
|
lumican
|
|
chr11_+_67013553
|
4.220
|
NM_030679
|
Myh1
|
myosin, heavy polypeptide 1, skeletal muscle, adult
|
|
chr16_-_36334417
|
4.202
|
NM_001001332
|
BC117090
|
cDNA sequence BC1179090
|
|
chrX_+_98644459
|
4.192
|
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr11_+_68970573
|
4.187
|
NM_009659
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type
|
|
chr5_+_8056508
|
4.170
|
NM_001080974
|
Sri
|
sorcin
|
|
chr18_+_60535680
|
4.158
|
NM_001146275
NM_021792
|
Iigp1
|
interferon inducible GTPase 1
|
|
chr2_+_33071451
|
4.147
|
NM_011923
|
Angptl2
|
angiopoietin-like 2
|
|
chr16_-_10313924
|
4.145
|
|
Emp2
|
epithelial membrane protein 2
|
|
chr10_-_111434231
|
4.140
|
|
Glipr1
|
GLI pathogenesis-related 1 (glioma)
|
|
chr16_-_45844343
|
4.131
|
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr11_+_68970595
|
4.129
|
|
Alox12b
|
arachidonate 12-lipoxygenase, 12R type
|
|
chr10_+_92951804
|
4.128
|
|
Hal
|
histidine ammonia lyase
|
|
chr13_-_93564652
|
4.123
|
NM_011582
|
Thbs4
|
thrombospondin 4
|
|
chr1_+_133535033
|
4.106
|
|
Ctse
|
cathepsin E
|
|
chr3_-_88307024
|
4.095
|
|
Lmna
|
lamin A
|
|
chr11_-_54846882
|
4.072
|
NM_001110211
NM_013472
|
Anxa6
|
annexin A6
|
|
chr19_-_57290515
|
4.062
|
NM_178688
|
Ablim1
|
actin-binding LIM protein 1
|
|
chr13_-_74945368
|
4.039
|
NM_009817
|
Cast
|
calpastatin
|
|
chr8_-_109927080
|
4.038
|
NM_008706
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1
|
|
chr7_+_108534039
|
4.031
|
NM_027180
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1
|
|
chr11_-_20731016
|
4.028
|
NM_173752
|
1110067D22Rik
|
RIKEN cDNA 1110067D22 gene
|
|
chr3_-_92568947
|
4.021
|
NM_026335
|
Lce1h
|
late cornified envelope 1H
|
|
chr8_+_83017933
|
3.989
|
NM_010369
|
Gypa
|
glycophorin A
|
|
chr1_+_78813482
|
3.982
|
NM_021342
|
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4
|
|
chr6_+_29409404
|
3.978
|
|
Flnc
|
filamin C, gamma
|
|
chr1_-_132359292
|
3.971
|
NM_010016
|
Cd55
|
CD55 antigen
|
|
chr19_-_11340591
|
3.967
|
NM_007641
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1
|
|
chr3_-_142058569
|
3.962
|
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr19_-_9162377
|
3.962
|
NM_011681
|
Scgb1a1
|
secretoglobin, family 1A, member 1 (uteroglobin)
|
|
chr16_+_49855786
|
3.957
|
|
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer)
|
|
chr6_+_35202672
|
3.917
|
|
1810058I24Rik
|
RIKEN cDNA 1810058I24 gene
|
|
chr10_+_41196119
|
3.916
|
NM_001164433
NM_138315
|
Mical1
|
microtubule associated monoxygenase, calponin and LIM domain containing 1
|
|
chr5_+_93696645
|
3.890
|
|
Ccng2
|
cyclin G2
|
|
chr15_-_67008433
|
3.887
|
NM_009177
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr7_+_149628285
|
3.882
|
NM_009405
|
Tnni2
|
troponin I, skeletal, fast 2
|
|
chr12_+_112680868
|
3.863
|
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr7_+_3655919
|
3.851
|
|
Rps9
|
ribosomal protein S9
|
|
chrX_+_93291377
|
3.846
|
NM_010833
|
Msn
|
moesin
|
|
chr17_+_35331446
|
3.796
|
NM_008518
|
Ltb
|
lymphotoxin B
|
|
chrX_+_33428784
|
3.793
|
NM_001009947
|
Dock11
|
dedicator of cytokinesis 11
|
|
chr13_+_3553690
|
3.790
|
|
Gdi2
|
guanosine diphosphate (GDP) dissociation inhibitor 2
|
|
chr5_+_93696566
|
3.755
|
NM_007635
|
Ccng2
|
cyclin G2
|
|
chr2_-_140497116
|
3.748
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_-_115758729
|
3.745
|
NM_172086
|
Rpl32
|
ribosomal protein L32
|
|
chr9_-_71440061
|
3.741
|
NM_001033208
|
Gcom1
|
GRINL1A complex locus
|
|
chr11_+_43287820
|
3.739
|
NM_026979
|
C1qtnf2
|
C1q and tumor necrosis factor related protein 2
|
|
chr8_+_70018744
|
3.738
|
NM_001168577
NM_010874
|
Nat2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase)
|
|
chr19_-_5726221
|
3.737
|
NM_001114595
NM_001114596
NM_001114597
NM_053252
|
Ehbp1l1
|
EH domain binding protein 1-like 1
|
|
chr10_+_115254308
|
3.722
|
NM_001168679
NM_001168680
NM_146010
|
Tspan8
|
tetraspanin 8
|
|
chr15_-_34372832
|
3.717
|
NM_009083
NM_001163485
|
Rpl30
|
ribosomal protein L30
|
|
chr16_+_96422285
|
3.716
|
|
Sh3bgr
|
SH3-binding domain glutamic acid-rich protein
|
|
chr3_-_36374572
|
3.715
|
|
Anxa5
|
annexin A5
|
|
chr15_-_5193681
|
3.691
|
NM_001136079
NM_008965
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr5_-_138277390
|
3.684
|
|
Pilra
|
paired immunoglobin-like type 2 receptor alpha
|
|
chr11_+_43287798
|
3.673
|
|
C1qtnf2
|
C1q and tumor necrosis factor related protein 2
|
|
chr2_-_119980210
|
3.672
|
|
Ehd4
|
EH-domain containing 4
|
|
chrX_+_98644445
|
3.667
|
NM_013712
|
Itgb1bp2
|
integrin beta 1 binding protein 2
|
|
chr15_+_78813524
|
3.662
|
|
Triobp
|
TRIO and F-actin binding protein
|
|
chr17_-_7997838
|
3.649
|
NM_001081416
|
Fndc1
|
fibronectin type III domain containing 1
|
|
chr12_+_8778152
|
3.647
|
|
Sdc1
|
syndecan 1
|
|
chr4_+_118100682
|
3.631
|
NM_001039175
NM_019422
|
Elovl1
|
elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
|
|
chr17_-_57367542
|
3.610
|
NM_009778
|
C3
|
complement component 3
|
|
chr3_-_142058595
|
3.609
|
NM_001190852
NM_001190853
NM_001190854
NM_001190855
NM_001190856
NM_019808
NM_019809
|
Pdlim5
|
PDZ and LIM domain 5
|
|
chr17_-_45730551
|
3.609
|
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr18_+_20469364
|
3.596
|
NM_010079
|
Dsg1a
|
desmoglein 1 alpha
|
|
chr9_+_46036788
|
3.590
|
|
Apoa1
|
apolipoprotein A-I
|
|
chr15_+_81977766
|
3.587
|
|
Srebf2
|
sterol regulatory element binding factor 2
|
|
chr16_+_14163414
|
3.571
|
|
Nde1
|
nuclear distribution gene E homolog 1 (A nidulans)
|
|
chr12_+_112680869
|
3.571
|
NM_009396
|
Tnfaip2
|
tumor necrosis factor, alpha-induced protein 2
|
|
chr2_-_140497204
|
3.569
|
NM_001172160
NM_178382
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr19_-_4190836
|
3.565
|
NM_178650
|
Tbc1d10c
|
TBC1 domain family, member 10c
|
|
chr9_-_110649625
|
3.556
|
NM_011199
|
Pth1r
|
parathyroid hormone 1 receptor
|
|
chr3_-_36374562
|
3.555
|
|
Anxa5
|
annexin A5
|
|
chr7_+_3293008
|
3.545
|
NM_001093766
|
Myadm
|
myeloid-associated differentiation marker
|
|
chr7_+_50663608
|
3.541
|
NM_172900
|
Siglecg
|
sialic acid binding Ig-like lectin G
|
|
chr11_+_109512220
|
3.502
|
NM_021880
|
Prkar1a
|
protein kinase, cAMP dependent regulatory, type I, alpha
|
|
chr2_-_140497183
|
3.499
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr6_-_87285690
|
3.489
|
NM_054041
|
Antxr1
|
anthrax toxin receptor 1
|
|
chr17_+_35576081
|
3.473
|
NM_001198560
NM_001198561
NM_010394
|
H2-Q9
H2-Q7
|
histocompatibility 2, Q region locus 9
histocompatibility 2, Q region locus 7
|
|
chr7_-_141416778
|
3.469
|
NM_007400
|
Adam12
|
a disintegrin and metallopeptidase domain 12 (meltrin alpha)
|
|
chr17_-_45724536
|
3.469
|
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1
|
|
chr9_-_45012097
|
3.456
|
NM_145403
|
Tmprss4
|
transmembrane protease, serine 4
|
|
chr2_-_73367455
|
3.436
|
NM_153138
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr3_-_36374517
|
3.433
|
|
Anxa5
|
annexin A5
|
|
chr3_+_159502646
|
3.431
|
NM_026582
|
Wls
|
wntless homolog (Drosophila)
|
|
chr5_-_17341534
|
3.429
|
|
Cd36
|
CD36 antigen
|
|
chr9_+_46036703
|
3.418
|
NM_009692
|
Apoa1
|
apolipoprotein A-I
|
|
chr14_-_56407622
|
3.411
|
NM_001164107
NM_001164108
NM_019955
|
Ripk3
|
receptor-interacting serine-threonine kinase 3
|
|
chrX_+_71516128
|
3.388
|
NM_052835
|
Rpl10
|
ribosomal protein 10
|