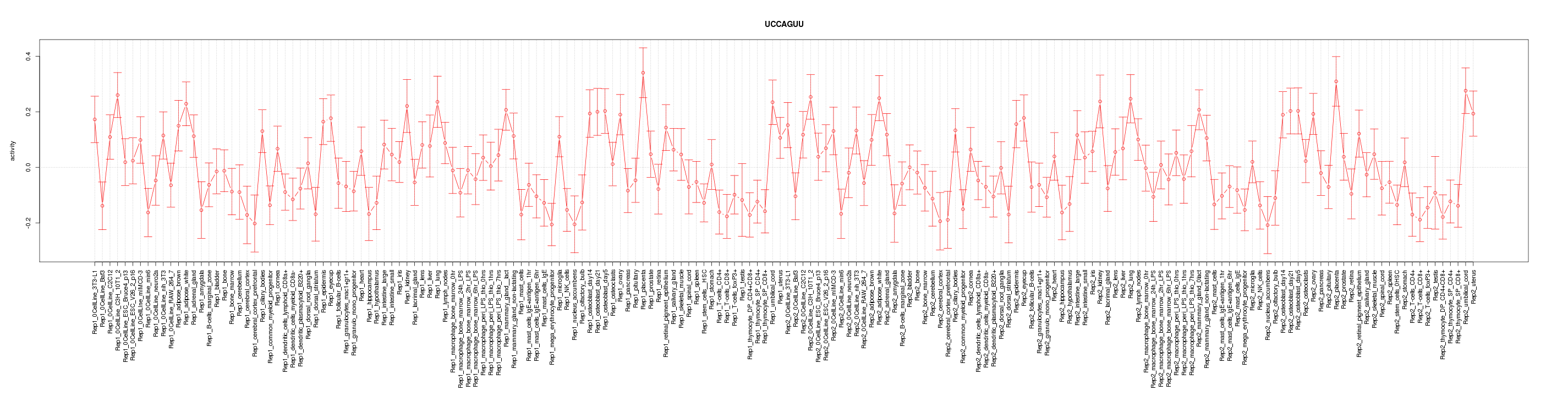
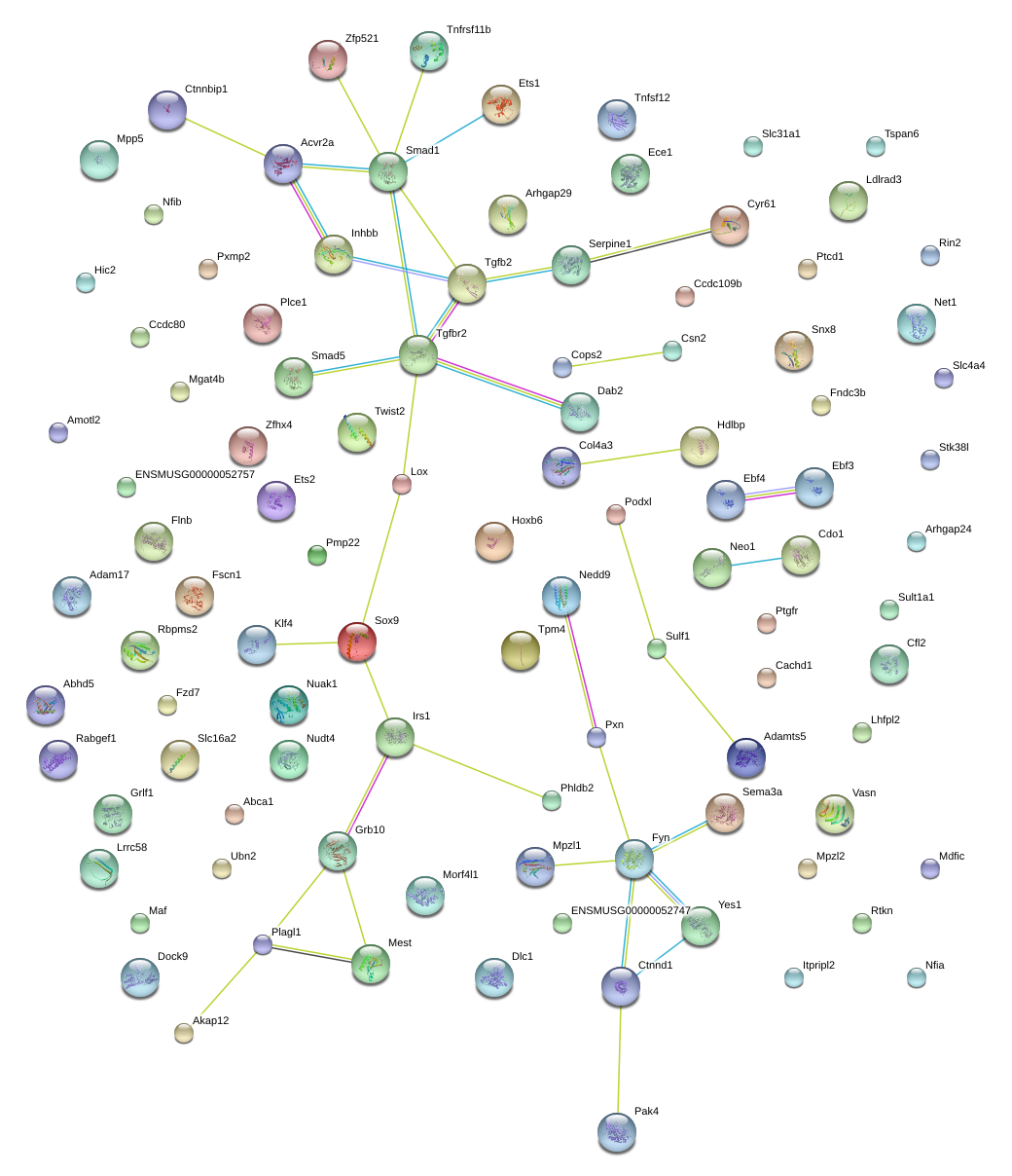
|
chr15_+_6249788
|
31.239
|
NM_001008702
|
Dab2
|
disabled homolog 2 (Drosophila)
|
|
chr16_+_4639921
|
28.602
|
NM_139307
|
Vasn
|
vasorin
|
|
chr15_+_6336747
|
25.058
|
NM_001037905
NM_001102400
NM_023118
|
Dab2
|
disabled homolog 2 (Drosophila)
|
|
chr1_-_82287990
|
24.857
|
NM_010570
|
Irs1
|
insulin receptor substrate 1
|
|
chr3_-_145312928
|
22.673
|
NM_010516
|
Cyr61
|
cysteine rich protein 61
|
|
chr4_-_55545337
|
22.364
|
NM_010637
|
Klf4
|
Kruppel-like factor 4 (gut)
|
|
chr11_-_11927973
|
21.952
|
NM_001177629
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr13_-_3892788
|
21.449
|
NM_001047159
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr16_-_85901362
|
21.044
|
NM_011782
|
Adamts5
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 5 (aggrecanase-2)
|
|
chr11_-_11937330
|
20.688
|
NM_010345
|
Grb10
|
growth factor receptor bound protein 10
|
|
chr11_+_62944973
|
20.008
|
NM_008885
|
Pmp22
|
peripheral myelin protein 22
|
|
chr13_-_3917356
|
18.572
|
NM_019671
|
Net1
|
neuroepithelial cell transforming gene 1
|
|
chr6_+_15670660
|
17.599
|
NM_175088
|
Mdfic
|
MyoD family inhibitor domain containing
|
|
chr10_-_6080145
|
17.485
|
NM_031185
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12
|
|
chr10_+_12810593
|
16.337
|
NM_009538
|
Plagl1
|
pleiomorphic adenoma gene-like 1
|
|
chr6_+_30688050
|
16.066
|
NM_008590
|
Mest
|
mesoderm specific transcript
|
|
chr2_-_84490827
|
16.046
|
NM_001085448
NM_001085449
NM_001085450
NM_007615
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chr2_-_84490666
|
15.714
|
NM_001085453
|
Ctnnd1
|
catenin (cadherin associated protein), delta 1
|
|
chrX_-_130432840
|
15.242
|
NM_019656
|
Tspan6
|
tetraspanin 6
|
|
chr1_-_121318824
|
15.112
|
NM_008381
|
Inhbb
|
inhibin beta-B
|
|
chr4_-_82151180
|
14.941
|
NM_001113209
NM_001113210
NM_008687
|
Nfib
|
nuclear factor I/B
|
|
chr8_+_74659107
|
14.889
|
NM_001001491
|
Tpm4
|
tropomyosin 4
|
|
chr9_+_44850426
|
14.684
|
NM_007962
|
Mpzl2
|
myelin protein zero-like 2
|
|
chr8_-_37796104
|
14.659
|
NM_001194941
|
Dlc1
|
deleted in liver cancer 1
|
|
chr10_-_95026787
|
14.655
|
NM_027722
|
Nudt4
|
nudix (nucleoside diphosphate linked moiety X)-type motif 4
|
|
chr9_-_116084395
|
14.284
|
NM_009371
NM_029575
|
Tgfbr2
|
transforming growth factor, beta receptor II
|
|
chr8_-_38001686
|
14.230
|
NM_001194940
|
Dlc1
|
deleted in liver cancer 1
|
|
chr2_+_145611851
|
14.152
|
NM_028724
|
Rin2
|
Ras and Rab interactor 2
|
|
chr18_-_52689279
|
13.971
|
NM_010728
|
Lox
|
lysyl oxidase
|
|
chr19_+_38598686
|
13.772
|
NM_019588
|
Plce1
|
phospholipase C, epsilon 1
|
|
chr5_-_137548123
|
13.528
|
NM_008871
|
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1
|
|
chr1_+_93698037
|
13.057
|
NM_007855
|
Twist2
|
twist homolog 2 (Drosophila)
|
|
chr15_-_54109855
|
12.178
|
NM_008764
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin)
|
|
chr1_-_188529867
|
12.123
|
NM_009367
|
Tgfb2
|
transforming growth factor, beta 2
|
|
chr16_-_45844409
|
11.724
|
NM_153412
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2
|
|
chr7_-_133819870
|
11.703
|
NM_133670
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1
|
|
chr3_+_121656136
|
11.661
|
NM_172525
|
Arhgap29
|
Rho GTPase activating protein 29
|
|
chr1_+_12682510
|
11.640
|
NM_001198565
NM_001198566
|
Sulf1
|
sulfatase 1
|
|
chr10_-_83903215
|
11.413
|
NM_001004363
|
Nuak1
|
NUAK family, SNF1-like kinase, 1
|
|
chr1_+_12708582
|
11.200
|
NM_172294
|
Sulf1
|
sulfatase 1
|
|
chr8_-_118230771
|
10.971
|
NM_001025577
|
Maf
|
avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog
|
|
chr5_+_32913539
|
10.811
|
NM_009535
|
Yes1
|
Yamaguchi sarcoma viral (v-yes) oncogene homolog 1
|
|
chr9_+_65478364
|
10.715
|
NM_028030
|
Rbpms2
|
RNA binding protein with multiple splicing 2
|
|
chr5_+_102910409
|
10.567
|
NM_029270
NM_146161
|
Arhgap24
|
Rho GTPase activating protein 24
|
|
chr11_+_112643501
|
10.383
|
NM_011448
|
Sox9
|
SRY-box containing gene 9
|
|
chr8_-_37676995
|
9.961
|
NM_015802
|
Dlc1
|
deleted in liver cancer 1
|
|
chr4_+_97444312
|
9.798
|
NM_001122953
NM_010905
|
Nfia
|
nuclear factor I/A
|
|
chr18_-_14131046
|
9.798
|
NM_145492
|
Zfp521
|
zinc finger protein 521
|
|
chr13_+_94827750
|
9.596
|
NM_172589
|
Lhfpl2
|
lipoma HMGIC fusion partner-like 2
|
|
chr16_+_95923988
|
9.532
|
NM_011809
|
Ets2
|
E26 avian leukemia oncogene 2, 3' domain
|
|
chr4_+_62021728
|
9.382
|
NM_175090
|
Slc31a1
|
solute carrier family 31, member 1
|
|
chr4_+_97248556
|
9.346
|
NM_001122952
|
Nfia
D130039L10Rik
|
nuclear factor I/A
RIKEN cDNA D130039L10 gene
|
|
chr6_+_38383918
|
9.346
|
NM_177185
|
Ubn2
|
ubinuclein 2
|
|
chr6_-_31513817
|
9.286
|
NM_013723
|
Podxl
|
podocalyxin-like
|
|
chr5_+_143722032
|
9.184
|
NM_007984
|
Fscn1
|
fascin homolog 1, actin bundling protein (Strongylocentrotus purpuratus)
|
|
chr3_+_5218545
|
9.083
|
NM_030708
|
Zfhx4
|
zinc finger homeodomain 4
|
|
chr1_-_167564449
|
8.967
|
NM_001001880
NM_001083897
|
Mpzl1
|
myelin protein zero-like 1
|
|
chr5_+_13399301
|
8.778
|
NM_009152
|
Sema3a
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A
|
|
chr1_-_95375372
|
8.672
|
NM_133808
|
Hdlbp
|
high density lipoprotein (HDL) binding protein
|
|
chr6_+_146673451
|
8.542
|
NM_172734
|
Stk38l
|
serine/threonine kinase 38 like
|
|
chr11_-_69499014
|
8.530
|
NM_001159505
NM_023517
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13
|
|
chr6_+_83085799
|
8.526
|
NM_001136227
|
Rtkn
|
rhotekin
|
|
chr5_+_115956682
|
8.227
|
NM_011223
NM_133915
|
Pxn
|
paxillin
|
|
chr16_+_17233664
|
8.041
|
NM_178922
|
Hic2
|
hypermethylated in cancer 2
|
|
chr5_-_88128419
|
7.997
|
NM_009972
|
Csn2
|
casein beta
|
|
chr7_-_125635435
|
7.893
|
NM_001033380
|
Itpripl2
|
inositol 1,4,5-triphosphate receptor interacting protein-like 2
|
|
chr2_+_48669628
|
7.766
|
NM_007396
|
Acvr2a
|
activin receptor IIA
|
|
chr3_-_27609279
|
7.755
|
NM_173182
|
Fndc3b
|
fibronectin type III domain containing 3B
|
|
chr16_+_37868418
|
7.590
|
NM_177093
|
Lrrc58
|
leucine rich repeat containing 58
|
|
chr1_+_59538966
|
7.585
|
NM_008057
|
Fzd7
|
frizzled homolog 7 (Drosophila)
|
|
chr7_-_144506112
|
7.550
|
NM_001113414
NM_001113415
NM_010096
|
Ebf3
|
early B-cell factor 3
|
|
chr16_+_45094163
|
7.457
|
NM_026439
|
Ccdc80
|
coiled-coil domain containing 80
|
|
chr9_+_102620096
|
7.340
|
NM_019764
|
Amotl2
|
angiomotin-like 2
|
|
chr12_+_79849918
|
7.193
|
NM_019579
|
Mpp5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5)
|
|
chr18_-_46887869
|
7.186
|
NM_033037
|
Cdo1
|
cysteine dioxygenase 1, cytosolic
|
|
chr14_+_8650470
|
7.144
|
NM_134080
|
Flnb
|
filamin, beta
|
|
chr4_+_100449274
|
7.097
|
NM_198037
|
Cachd1
|
cache domain containing 1
|
|
chr5_+_89316245
|
7.046
|
NM_001136260
NM_001197147
NM_018760
|
Slc4a4
|
solute carrier family 4 (anion exchanger), member 4
|
|
chr7_-_17200285
|
7.029
|
NM_172739
|
Grlf1
|
glucocorticoid receptor DNA binding factor 1
|
|
chr3_-_129672992
|
6.519
|
NM_025779
|
Ccdc109b
|
coiled-coil domain containing 109B
|
|
chr4_+_137418151
|
6.499
|
NM_199307
|
Ece1
|
endothelin converting enzyme 1
|
|
chr11_-_69509498
|
6.446
|
NM_011614
NM_001034097
NM_001159503
|
Tnfsf12
BC096441
|
tumor necrosis factor (ligand) superfamily, member 12
cDNA sequence BC096441
|
|
chr13_+_56805028
|
6.334
|
NM_001164042
|
Smad5
|
MAD homolog 5 (Drosophila)
|
|
chr11_+_50038815
|
6.314
|
NM_145926
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B
|
|
chr9_+_122260733
|
6.276
|
NM_026179
|
Abhd5
|
abhydrolase domain containing 5
|
|
chr3_-_151500395
|
6.139
|
NM_008966
|
Ptgfr
|
prostaglandin F receptor
|
|
chrX_-_101017286
|
6.133
|
NM_009197
|
Slc16a2
|
solute carrier family 16 (monocarboxylic acid transporters), member 2
|
|
chr4_+_148892231
|
6.080
|
NM_023465
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr5_-_140865121
|
6.079
|
NM_172277
|
Snx8
|
sorting nexin 8
|
|
chr1_+_82583495
|
6.070
|
NM_007734
|
Col4a3
|
collagen, type IV, alpha 3
|
|
chr11_+_96160484
|
6.013
|
NM_008269
|
Hoxb6
|
homeobox B6
|
|
chr13_-_41582649
|
6.000
|
NM_001111324
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr7_-_29383180
|
5.915
|
NM_027470
|
Pak4
|
p21 protein (Cdc42/Rac)-activated kinase 4
|
|
chr4_-_53172748
|
5.913
|
NM_013454
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1
|
|
chr12_-_21379292
|
5.869
|
NM_009615
|
Adam17
|
a disintegrin and metallopeptidase domain 17
|
|
chr4_+_148919218
|
5.852
|
NM_001141930
|
Ctnnbip1
|
catenin beta interacting protein 1
|
|
chr12_-_55963776
|
5.714
|
NM_007688
|
Cfl2
|
cofilin 2, muscle
|
|
chr13_+_56804370
|
5.686
|
NM_008541
NM_001164041
|
Smad5
|
MAD homolog 5 (Drosophila)
|
|
chr2_-_102026486
|
5.575
|
NM_178886
|
Ldlrad3
|
low density lipoprotein receptor class A domain containing 3
|
|
chr14_-_122138082
|
5.572
|
NM_001128307
NM_001128308
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr14_-_122097583
|
5.557
|
NM_001081039
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr9_-_58884114
|
5.425
|
NM_001042752
NM_008684
|
Neo1
|
neogenin
|
|
chr1_-_24107142
|
5.380
|
NM_026604
|
Fam135a
|
family with sequence similarity 135, member A
|
|
chr7_+_75097142
|
5.378
|
NM_010513
|
Igf1r
|
insulin-like growth factor I receptor
|
|
chr10_+_17443024
|
5.302
|
NM_010828
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2
|
|
chr11_+_115243203
|
5.302
|
NM_001080929
|
Cdr2l
|
cerebellar degeneration-related protein 2-like
|
|
chr5_-_136410488
|
5.132
|
NM_018871
|
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide
|
|
chr18_+_83084023
|
4.970
|
NM_001177464
|
Zfp516
|
zinc finger protein 516
|
|
chr1_+_196446006
|
4.932
|
NM_008882
|
Plxna2
|
plexin A2
|
|
chr15_-_58654964
|
4.897
|
NM_175212
|
Tmem65
|
transmembrane protein 65
|
|
chr4_-_115840142
|
4.866
|
NM_133681
|
Tspan1
|
tetraspanin 1
|
|
chr18_-_16967557
|
4.762
|
NM_007664
|
Cdh2
|
cadherin 2
|
|
chr1_+_39957733
|
4.759
|
NM_008696
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr6_+_83087016
|
4.677
|
NM_009106
NM_133641
|
Rtkn
|
rhotekin
|
|
chr19_+_53751795
|
4.666
|
NM_001170847
|
Rbm20
|
RNA binding motif protein 20
|
|
chr1_+_174430359
|
4.547
|
NM_178598
|
Tagln2
|
transgelin 2
|
|
chr5_-_125821418
|
4.507
|
NM_016741
|
Scarb1
|
scavenger receptor class B, member 1
|
|
chr19_-_44144244
|
4.476
|
NM_001164359
NM_001164360
|
Erlin1
|
ER lipid raft associated 1
|
|
chr16_+_17452048
|
4.474
|
NM_007764
|
Crkl
|
v-crk sarcoma virus CT10 oncogene homolog (avian)-like
|
|
chr7_-_35070627
|
4.459
|
NM_172741
|
4931406P16Rik
|
RIKEN cDNA 4931406P16 gene
|
|
chr13_-_41454358
|
4.446
|
NM_017464
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9
|
|
chr2_-_104656852
|
4.358
|
NM_001123327
|
Qser1
|
glutamine and serine rich 1
|
|
chr3_-_84024766
|
4.251
|
NM_030706
|
Trim2
|
tripartite motif-containing 2
|
|
chr9_-_101154160
|
4.206
|
NM_001161362
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr5_+_28397902
|
4.128
|
NM_153526
|
Insig1
|
insulin induced gene 1
|
|
chr17_+_78599556
|
4.098
|
NM_015800
|
Crim1
|
cysteine rich transmembrane BMP regulator 1 (chordin like)
|
|
chr2_+_152987036
|
4.064
|
NM_133847
|
Tm9sf4
|
transmembrane 9 superfamily protein member 4
|
|
chr10_-_126558207
|
4.006
|
NM_001171026
NM_177614
|
Os9
|
amplified in osteosarcoma
|
|
chr6_+_100783598
|
3.993
|
NM_182939
|
Ppp4r2
|
protein phosphatase 4, regulatory subunit 2
|
|
chr11_+_83116148
|
3.990
|
NM_001035854
NM_027915
|
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit
|
|
chr9_-_63605745
|
3.967
|
NM_016769
|
Smad3
|
MAD homolog 3 (Drosophila)
|
|
chr17_-_71351775
|
3.789
|
NM_026064
|
2900073G15Rik
|
RIKEN cDNA 2900073G15 gene
|
|
chr7_-_26035683
|
3.654
|
NM_010155
|
Erf
|
Ets2 repressor factor
|
|
chr9_-_101101334
|
3.647
|
NM_172144
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha
|
|
chr17_-_68353397
|
3.607
|
NM_172964
|
Arhgap28
|
Rho GTPase activating protein 28
|
|
chr2_+_25258568
|
3.604
|
NM_008721
|
Npdc1
|
neural proliferation, differentiation and control gene 1
|
|
chr11_-_96690784
|
3.579
|
NM_001130450
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1
|
|
chr8_-_90483392
|
3.538
|
NM_033327
|
Zfp423
|
zinc finger protein 423
|
|
chr11_-_97049185
|
3.537
|
NM_008379
|
Kpnb1
|
karyopherin (importin) beta 1
|
|
chrX_-_138825032
|
3.534
|
NM_001033600
NM_019477
NM_207625
|
Acsl4
|
acyl-CoA synthetase long-chain family member 4
|
|
chr10_-_13108775
|
3.499
|
NM_001195066
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr6_-_127100954
|
3.498
|
NM_009829
|
Ccnd2
|
cyclin D2
|
|
chr16_+_94791713
|
3.495
|
NM_007890
|
Dyrk1a
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1a
|
|
chr3_+_126299858
|
3.481
|
NM_001025438
NM_001025439
NM_023813
|
Camk2d
|
calcium/calmodulin-dependent protein kinase II, delta
|
|
chr14_-_122196929
|
3.454
|
NM_134074
|
Dock9
|
dedicator of cytokinesis 9
|
|
chr11_-_96691261
|
3.446
|
NM_008686
|
Nfe2l1
|
nuclear factor, erythroid derived 2,-like 1
|
|
chr5_-_86601756
|
3.442
|
NM_172712
|
Uba6
|
ubiquitin-like modifier activating enzyme 6
|
|
chr2_-_104334610
|
3.387
|
NM_001145824
NM_010434
|
Hipk3
|
homeodomain interacting protein kinase 3
|
|
chr9_+_106356125
|
3.373
|
NM_021567
|
Pcbp4
|
poly(rC) binding protein 4
|
|
chr5_+_139676589
|
3.320
|
NM_024451
|
Sun1
|
Sad1 and UNC84 domain containing 1
|
|
chr11_-_70832737
|
3.221
|
NM_033562
|
Derl2
|
Der1-like domain family, member 2
|
|
chr15_+_40486577
|
3.176
|
NM_011766
|
Zfpm2
|
zinc finger protein, multitype 2
|
|
chr3_-_142544883
|
3.170
|
NM_178654
|
Pkn2
|
protein kinase N2
|
|
chr9_-_21602942
|
3.131
|
NM_145611
|
Kank2
|
KN motif and ankyrin repeat domains 2
|
|
chr19_+_38911056
|
3.100
|
NM_145952
|
Tbc1d12
|
TBC1D12: TBC1 domain family, member 12
|
|
chr9_-_79566271
|
2.997
|
NM_007730
|
Col12a1
|
collagen, type XII, alpha 1
|
|
chr6_-_115944822
|
2.993
|
NM_026376
|
Plxnd1
|
plexin D1
|
|
chr13_-_108680257
|
2.832
|
NM_145456
|
Zswim6
|
zinc finger, SWIM domain containing 6
|
|
chr9_+_74800828
|
2.727
|
NM_001113283
|
BC031353
|
cDNA sequence BC031353
|
|
chr7_-_117758413
|
2.693
|
NM_177324
|
Sbf2
|
SET binding factor 2
|
|
chr9_-_110379093
|
2.657
|
NM_177771
|
Klhl18
|
kelch-like 18 (Drosophila)
|
|
chr1_+_36125177
|
2.652
|
NM_015818
|
Hs6st1
|
heparan sulfate 6-O-sulfotransferase 1
|
|
chr4_+_129497720
|
2.598
|
NM_008974
|
Ptp4a2
|
protein tyrosine phosphatase 4a2
|
|
chr7_-_91300359
|
2.513
|
NM_133722
|
Fam108c
|
family with sequence similarity 108, member C
|
|
chr10_-_13043959
|
2.489
|
NM_001033257
NM_001195096
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr18_+_69505374
|
2.459
|
NM_001083967
|
Tcf4
|
transcription factor 4
|
|
chr11_+_45869473
|
2.458
|
NM_009616
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta)
|
|
chrX_-_101176600
|
2.447
|
NM_011276
|
Rlim
|
ring finger protein, LIM domain interacting
|
|
chr6_-_39156706
|
2.419
|
NM_001033430
|
Jhdm1d
|
jumonji C domain-containing histone demethylase 1 homolog D (S. cerevisiae)
|
|
chr4_+_107107195
|
2.391
|
NM_147221
|
Glis1
|
GLIS family zinc finger 1
|
|
chr1_+_182656137
|
2.220
|
NM_133225
|
Acbd3
|
acyl-Coenzyme A binding domain containing 3
|
|
chr8_-_35040277
|
2.127
|
NM_001042674
NM_001042675
NM_019733
|
Rbpms
|
RNA binding protein gene with multiple splicing
|
|
chr8_-_66684789
|
2.092
|
NM_009390
|
Tll1
|
tolloid-like
|
|
chr11_-_18918923
|
2.063
|
NM_001193271
NM_010789
|
Meis1
|
Meis homeobox 1
|
|
chr10_-_13194201
|
2.043
|
NM_001195065
|
Phactr2
|
phosphatase and actin regulator 2
|
|
chr14_+_59820064
|
2.033
|
NM_027764
|
Rcbtb1
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1
|
|
chr18_+_69504145
|
2.028
|
NM_013685
|
Tcf4
|
transcription factor 4
|
|
chr13_+_47289371
|
1.969
|
NM_001170643
|
Rnf144b
|
ring finger protein 144B
|
|
chr13_+_119503505
|
1.957
|
NM_008002
|
Fgf10
|
fibroblast growth factor 10
|
|
chr5_+_122161617
|
1.957
|
NM_009125
|
Atxn2
|
ataxin 2
|
|
chr2_-_150835067
|
1.956
|
NM_207204
|
Ninl
|
ninein-like
|
|
chr9_-_35007875
|
1.952
|
NM_001177845
|
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein
|
|
chr13_+_37980739
|
1.915
|
NM_001177869
|
Rreb1
|
ras responsive element binding protein 1
|
|
chr1_+_44176812
|
1.911
|
NM_144558
|
Bivm
|
basic, immunoglobulin-like variable motif containing
|
|
chr12_+_70473032
|
1.907
|
NM_007481
|
Arf6
|
ADP-ribosylation factor 6
|
|
chr11_+_115626733
|
1.872
|
NM_028014
|
2310067B10Rik
|
RIKEN cDNA 2310067B10 gene
|
|
chr9_-_67407509
|
1.868
|
NM_001081242
|
Tln2
|
talin 2
|
|
chr16_+_14361637
|
1.803
|
NM_008576
|
Abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1
|
|
chr9_-_35007458
|
1.802
|
NM_001177847
NM_001177846
NM_054096
|
Tirap
|
toll-interleukin 1 receptor (TIR) domain-containing adaptor protein
|
|
chr9_+_53698053
|
1.765
|
NM_025540
|
Sln
|
sarcolipin
|
|
chr2_-_103637096
|
1.713
|
NM_001111289
NM_001111290
NM_001111292
|
Caprin1
|
cell cycle associated protein 1
|
|
chr9_-_59334089
|
1.707
|
NM_019927
|
Arih1
|
ariadne ubiquitin-conjugating enzyme E2 binding protein homolog 1 (Drosophila)
|
|
chr3_+_104442568
|
1.703
|
NM_009196
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1
|
|
chr15_-_27955602
|
1.683
|
NM_001081302
|
Trio
|
triple functional domain (PTPRF interacting)
|
|
chr18_-_63851841
|
1.682
|
NM_016792
|
Txnl1
|
thioredoxin-like 1
|
|
chrX_-_108651553
|
1.659
|
NM_025949
|
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6
|
|
chr9_-_97011440
|
1.633
|
NM_138756
|
Slc25a36
|
solute carrier family 25, member 36
|
|
chr4_+_106584074
|
1.630
|
NM_023672
NM_198438
|
Ssbp3
|
single-stranded DNA binding protein 3
|
|
chr7_+_4088622
|
1.625
|
NM_172736
|
Leng8
|
leukocyte receptor cluster (LRC) member 8
|
|
chr6_+_83187368
|
1.588
|
NM_001166067
|
Slc4a5
|
solute carrier family 4, sodium bicarbonate cotransporter, member 5
|